Arenavirus variations due to host-specific adaptation
- PMID: 23344562
- PMCID: PMC3564120
- DOI: 10.3390/v5010241
Arenavirus variations due to host-specific adaptation
Abstract
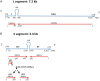
Arenavirus particles are enveloped and contain two single-strand RNA genomic segments with ambisense coding. Genetic plasticity of the arenaviruses comes from transcription errors, segment reassortment, and permissive genomic packaging, and results in their remarkable ability, as a group, to infect a wide variety of hosts. In this review, we discuss some in vitro studies of virus genetic and phenotypic variation after exposure to selective pressures such as high viral dose, mutagens and antivirals. Additionally, we discuss the variation in vivo of selected isolates of Old World arenaviruses, particularly after infection of different animal species. We also discuss the recent emergence of new arenaviruses in the context of our observations of sequence variations that appear to be host-specific.
Figures
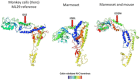
References
-
- Eichler R., Strecker T., Kolesnikova L., ter Meulen J., Weissenhorn W., Becker S., Klenk H.D., Garten W., Lenz O. Characterization of the lassa virus matrix protein Z: Electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP) Virus Res. 2004;100:249–255. doi: 10.1016/j.virusres.2003.11.017. - DOI - PubMed
-
- Casabona J.C., Levingston Macleod J.M., Loureiro M.E., Gomez G.A., Lopez N. The ring domain and the l79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J. Virol. 2009;83:7029–7039. doi: 10.1128/JVI.00329-09. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

