Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells
- PMID: 23344621
- PMCID: PMC3381593
- DOI: 10.1038/mtna.2011.3
Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells
Abstract
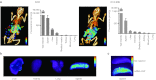
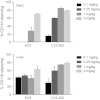
Leukocytes are central regulators of inflammation and the target cells of therapies for key diseases, including autoimmune, cardiovascular, and malignant disorders. Efficient in vivo delivery of small interfering RNA (siRNA) to immune cells could thus enable novel treatment strategies with broad applicability. In this report, we develop systemic delivery methods of siRNA encapsulated in lipid nanoparticles (LNP) for durable and potent in vivo RNA interference (RNAi)-mediated silencing in myeloid cells. This work provides the first demonstration of siRNA-mediated silencing in myeloid cell types of nonhuman primates (NHPs) and establishes the feasibility of targeting multiple gene targets in rodent myeloid cells. The therapeutic potential of these formulations was demonstrated using siRNA targeting tumor necrosis factor-α (TNFα) which induced substantial attenuation of disease progression comparable to a potent antibody treatment in a mouse model of rheumatoid arthritis (RA). In summary, we demonstrate a broadly applicable and therapeutically relevant platform for silencing disease genes in immune cells.
Figures




Similar articles
-
Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics.Acc Chem Res. 2019 Sep 17;52(9):2435-2444. doi: 10.1021/acs.accounts.9b00368. Epub 2019 Aug 9. Acc Chem Res. 2019. PMID: 31397996 Review.
-
Lipidoid-polymer hybrid nanoparticles loaded with TNF siRNA suppress inflammation after intra-articular administration in a murine experimental arthritis model.Eur J Pharm Biopharm. 2019 Sep;142:38-48. doi: 10.1016/j.ejpb.2019.06.009. Epub 2019 Jun 11. Eur J Pharm Biopharm. 2019. PMID: 31199978
-
Optimizing the Intracellular Delivery of Therapeutic Anti-inflammatory TNF-α siRNA to Activated Macrophages Using Lipidoid-Polymer Hybrid Nanoparticles.Front Bioeng Biotechnol. 2021 Jan 14;8:601155. doi: 10.3389/fbioe.2020.601155. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33520957 Free PMC article.
-
Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells.Mol Ther. 2011 Dec;19(12):2186-200. doi: 10.1038/mt.2011.190. Epub 2011 Oct 4. Mol Ther. 2011. PMID: 21971424 Free PMC article.
-
Lipid nanoparticles for short interfering RNA delivery.Adv Genet. 2014;88:71-110. doi: 10.1016/B978-0-12-800148-6.00004-3. Adv Genet. 2014. PMID: 25409604 Free PMC article. Review.
Cited by
-
Proof-of-concept Studies for siRNA-mediated Gene Silencing for Coagulation Factors in Rat and Rabbit.Mol Ther Nucleic Acids. 2015 Jan 27;4(1):e224. doi: 10.1038/mtna.2014.75. Mol Ther Nucleic Acids. 2015. PMID: 25625614 Free PMC article.
-
Nanoparticles and their applications in cell and molecular biology.Integr Biol (Camb). 2014 Jan;6(1):9-26. doi: 10.1039/c3ib40165k. Integr Biol (Camb). 2014. PMID: 24104563 Free PMC article. Review.
-
Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates.Proc Natl Acad Sci U S A. 2014 Mar 18;111(11):3955-60. doi: 10.1073/pnas.1322937111. Epub 2014 Feb 10. Proc Natl Acad Sci U S A. 2014. PMID: 24516150 Free PMC article.
-
Osteogenic effects of microRNA-335-5p/lipidoid nanoparticles coated on titanium surface.Arch Oral Biol. 2021 Sep;129:105207. doi: 10.1016/j.archoralbio.2021.105207. Epub 2021 Jul 6. Arch Oral Biol. 2021. PMID: 34273868 Free PMC article.
-
Macrophage Notch Ligand Delta-Like 4 Promotes Vein Graft Lesion Development: Implications for the Treatment of Vein Graft Failure.Arterioscler Thromb Vasc Biol. 2015 Nov;35(11):2343-2353. doi: 10.1161/ATVBAHA.115.305516. Epub 2015 Sep 24. Arterioscler Thromb Vasc Biol. 2015. PMID: 26404485 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

