Targeted exon skipping to address "leaky" mutations in the dystrophin gene
- PMID: 23344648
- PMCID: PMC3499695
- DOI: 10.1038/mtna.2012.40
Targeted exon skipping to address "leaky" mutations in the dystrophin gene
Abstract
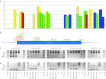
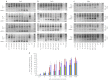
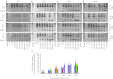
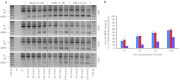
Protein-truncating mutations in the dystrophin gene lead to the progressive muscle wasting disorder Duchenne muscular dystrophy, whereas in-frame deletions typically manifest as the milder allelic condition, Becker muscular dystrophy. Antisense oligomer-induced exon skipping can modify dystrophin gene expression so that a disease-associated dystrophin pre-mRNA is processed into a Becker muscular dystrophy-like mature transcript. Despite genomic deletions that may encompass hundreds of kilobases of the gene, some dystrophin mutations appear "leaky", and low levels of high molecular weight, and presumably semi-functional, dystrophin are produced. A likely causative mechanism is endogenous exon skipping, and Duchenne individuals with higher baseline levels of dystrophin may respond more efficiently to the administration of splice-switching antisense oligomers. We optimized excision of exons 8 and 9 in normal human myoblasts, and evaluated several oligomers in cells from eight Duchenne muscular dystrophy patients with deletions in a known "leaky" region of the dystrophin gene. Inter-patient variation in response to antisense oligomer induced skipping in vitro appeared minimal. We describe oligomers targeting exon 8, that unequivocally increase dystrophin above baseline in vitro, and propose that patients with leaky mutations are ideally suited for participation in antisense oligomer mediated splice-switching clinical studies.Molecular Therapy - Nucleic Acids (2012) 1, e48; doi:10.1038/mtna.2012.40; published online 16 October 2012.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

