Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes
- PMID: 23344904
- PMCID: PMC3585612
- DOI: 10.1104/pp.112.210971
Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes
Abstract
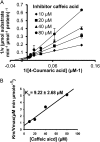
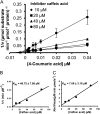
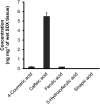
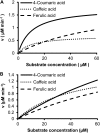
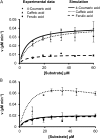
4-Coumaric acid:coenzyme A ligase (4CL) is involved in monolignol biosynthesis for lignification in plant cell walls. It ligates coenzyme A (CoA) with hydroxycinnamic acids, such as 4-coumaric and caffeic acids, into hydroxycinnamoyl-CoA thioesters. The ligation ensures the activated state of the acid for reduction into monolignols. In Populus spp., it has long been thought that one monolignol-specific 4CL is involved. Here, we present evidence of two monolignol 4CLs, Ptr4CL3 and Ptr4CL5, in Populus trichocarpa. Ptr4CL3 is the ortholog of the monolignol 4CL reported for many other species. Ptr4CL5 is novel. The two Ptr4CLs exhibited distinct Michaelis-Menten kinetic properties. Inhibition kinetics demonstrated that hydroxycinnamic acid substrates are also inhibitors of 4CL and suggested that Ptr4CL5 is an allosteric enzyme. Experimentally validated flux simulation, incorporating reaction/inhibition kinetics, suggested two CoA ligation paths in vivo: one through 4-coumaric acid and the other through caffeic acid. We previously showed that a membrane protein complex mediated the 3-hydroxylation of 4-coumaric acid to caffeic acid. The demonstration here of two ligation paths requiring these acids supports this 3-hydroxylation function. Ptr4CL3 regulates both CoA ligation paths with similar efficiencies, whereas Ptr4CL5 regulates primarily the caffeic acid path. Both paths can be inhibited by caffeic acid. The Ptr4CL5-catalyzed caffeic acid metabolism, therefore, may also act to mitigate the inhibition by caffeic acid to maintain a proper ligation flux. A high level of caffeic acid was detected in stem-differentiating xylem of P. trichocarpa. Our results suggest that Ptr4CL5 and caffeic acid coordinately modulate the CoA ligation flux for monolignol biosynthesis.
Figures

References
-
- Bolwell G, Cramer C, Lamb C, Schuch W, Dixon R. (1986) L-Phenylalanine ammonia-lyase from Phaseolus vulgaris: modulation of the levels of active enzyme by trans-cinnamic acid. Planta 169: 97–107 - PubMed
-
- Brown S, Neish A. (1959) Studies of lignin biosynthesis using isotopic carbon. VIII. Isolation of radioactive hydrogenolysis products of lignin1. J Am Chem Soc 81: 2419–2424
-
- Brown SA. (1961) Chemistry of lignification: biochemical research on lignins is yielding clues to the structure and formation of these complex polymers. Science 134: 305–313 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

