Pattern and synchrony of gene expression among sympatric marine microbial populations
- PMID: 23345438
- PMCID: PMC3568374
- DOI: 10.1073/pnas.1222099110
Pattern and synchrony of gene expression among sympatric marine microbial populations
Abstract
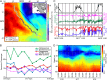
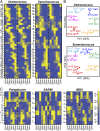
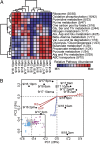
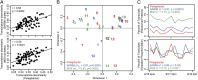
Planktonic marine microbes live in dynamic habitats that demand rapid sensing and response to periodic as well as stochastic environmental change. The kinetics, regularity, and specificity of microbial responses in situ, however, are not well-described. We report here simultaneous multitaxon genome-wide transcriptome profiling in a naturally occurring picoplankton community. An in situ robotic sampler using a Lagrangian sampling strategy enabled continuous tracking and repeated sampling of coherent microbial populations over 2 d. Subsequent RNA sequencing analyses yielded genome-wide transcriptome profiles of eukaryotic (Ostreococcus) and bacterial (Synechococcus) photosynthetic picoplankton as well as proteorhodopsin-containing heterotrophs, including Pelagibacter, SAR86-cluster Gammaproteobacteria, and marine Euryarchaea. The photosynthetic picoplankton exhibited strong diel rhythms over thousands of gene transcripts that were remarkably consistent with diel cycling observed in laboratory pure cultures. In contrast, the heterotrophs did not cycle diurnally. Instead, heterotrophic picoplankton populations exhibited cross-species synchronous, tightly regulated, temporally variable patterns of gene expression for many genes, particularly those genes associated with growth and nutrient acquisition. This multitaxon, population-wide gene regulation seemed to reflect sporadic, short-term, reversible responses to high-frequency environmental variability. Although the timing of the environmental responses among different heterotrophic species seemed synchronous, the specific metabolic genes that were expressed varied from taxon to taxon. In aggregate, these results provide insights into the kinetics, diversity, and functional patterns of microbial community response to environmental change. Our results also suggest a means by which complex multispecies metabolic processes could be coordinated, facilitating the regulation of matter and energy processing in a dynamically changing environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10(9):410–418. - PubMed
-
- Poretsky RS, et al. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11(6):1358–1375. - PubMed
-
- Hewson I, Poretsky RS, Tripp HJ, Montoya JP, Zehr JP. Spatial patterns and light-driven variation of microbial population gene expression in surface waters of the oligotrophic open ocean. Environ Microbiol. 2010;12(7):1940–1956. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

