Enhanced autointegration in hyperstable simian immunodeficiency virus capsid mutants blocked after reverse transcription
- PMID: 23345510
- PMCID: PMC3624228
- DOI: 10.1128/JVI.03239-12
Enhanced autointegration in hyperstable simian immunodeficiency virus capsid mutants blocked after reverse transcription
Abstract
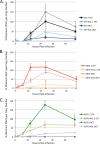
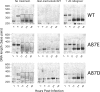
After entering a host cell, retroviruses such as simian immunodeficiency virus (SIV) uncoat, disassembling the viral capsid. Rates of uncoating that are too high and too low can be detrimental to the efficiency of infection. Rapid uncoating typically leads to blocks in reverse transcription, but the basis for replication defects associated with slow uncoating is less clear. Here we characterize the phenotypes of two SIVmac239 mutants with changes, A87E and A87D, in the helix 4/5 loop of the capsid protein. These mutant viruses exhibited normal capsid morphology but were significantly attenuated for infectivity. The infectivity of wild-type and mutant SIVmac239 was not decreased by aphidicolin-induced growth arrest of the target cells. In the cytosol of infected cells, the A87E and A87D capsids remained in particulate form longer than the wild-type SIVmac239 capsid, suggesting that the mutants uncoat more slowly than the wild-type capsid. Both mutants exhibited much higher levels of autointegrated DNA forms than wild-type SIVmac239. Thus, some changes in the helix 4/5 loop of the SIVmac239 capsid protein result in capsid hyperstability and an increase in autointegration.
Figures
Similar articles
-
Contribution of glutamine residues in the helix 4-5 loop to capsid-capsid interactions in simian immunodeficiency virus of macaques.J Virol. 2014 Sep;88(18):10289-302. doi: 10.1128/JVI.01388-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991000 Free PMC article.
-
Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus.J Virol. 2011 Aug;85(16):8116-32. doi: 10.1128/JVI.00341-11. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680520 Free PMC article.
-
Degradation of SAMHD1 by Vpx Is Independent of Uncoating.J Virol. 2015 May;89(10):5701-13. doi: 10.1128/JVI.03575-14. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762741 Free PMC article.
-
May I Help You with Your Coat? HIV-1 Capsid Uncoating and Reverse Transcription.Int J Mol Sci. 2024 Jun 28;25(13):7167. doi: 10.3390/ijms25137167. Int J Mol Sci. 2024. PMID: 39000271 Free PMC article. Review.
-
Disassembling the Nature of Capsid: Biochemical, Genetic, and Imaging Approaches to Assess HIV-1 Capsid Functions.Viruses. 2021 Nov 7;13(11):2237. doi: 10.3390/v13112237. Viruses. 2021. PMID: 34835043 Free PMC article. Review.
Cited by
-
The Integrase: An Overview of a Key Player Enzyme in the Antiviral Scenario.Int J Mol Sci. 2023 Jul 29;24(15):12187. doi: 10.3390/ijms241512187. Int J Mol Sci. 2023. PMID: 37569561 Free PMC article. Review.
-
A model for cofactor use during HIV-1 reverse transcription and nuclear entry.Curr Opin Virol. 2014 Feb;4(100):32-6. doi: 10.1016/j.coviro.2013.11.003. Epub 2014 Jan 14. Curr Opin Virol. 2014. PMID: 24525292 Free PMC article. Review.
-
HIV-1 capsid is involved in post-nuclear entry steps.Retrovirology. 2016 Apr 23;13:28. doi: 10.1186/s12977-016-0262-0. Retrovirology. 2016. PMID: 27107820 Free PMC article.
-
Retroviral DNA Integration.Chem Rev. 2016 Oct 26;116(20):12730-12757. doi: 10.1021/acs.chemrev.6b00125. Epub 2016 May 20. Chem Rev. 2016. PMID: 27198982 Free PMC article. Review.
-
Contribution of glutamine residues in the helix 4-5 loop to capsid-capsid interactions in simian immunodeficiency virus of macaques.J Virol. 2014 Sep;88(18):10289-302. doi: 10.1128/JVI.01388-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991000 Free PMC article.
References
-
- Coffin JM, Hughes SH, Varmus H. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY - PubMed
-
- Leschonsky B, Ludwig C, Bieler K, Wagner R. 2007. Capsid stability and replication of human immunodeficiency virus type 1 are influenced critically by charge and size of Gag residue 183. J. Gen. Virol. 88:207–216 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

