Mge1, a nucleotide exchange factor of Hsp70, acts as an oxidative sensor to regulate mitochondrial Hsp70 function
- PMID: 23345595
- PMCID: PMC3596242
- DOI: 10.1091/mbc.E12-10-0719
Mge1, a nucleotide exchange factor of Hsp70, acts as an oxidative sensor to regulate mitochondrial Hsp70 function
Abstract
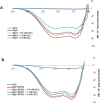
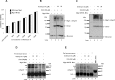
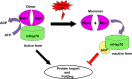
Despite the growing evidence of the role of oxidative stress in disease, its molecular mechanism of action remains poorly understood. The yeast Saccharomyces cerevisiae provides a valuable model system in which to elucidate the effects of oxidative stress on mitochondria in higher eukaryotes. Dimeric yeast Mge1, the cochaperone of heat shock protein 70 (Hsp70), is essential for exchanging ATP for ADP on Hsp70 and thus for recycling of Hsp70 for mitochondrial protein import and folding. Here we show an oxidative stress-dependent decrease in Mge1 dimer formation accompanied by a concomitant decrease in Mge1-Hsp70 complex formation in vitro. The Mge1-M155L substitution mutant stabilizes both Mge1 dimer and Mge1-Hsp70 complex formation. Most important, the Mge1-M155L mutant rescues the slow-growth phenomenon associated with the wild-type Mge1 strain in the presence of H2O2 in vivo, stimulation of the ATPase activity of Hsp70, and the protein import defect during oxidative stress in vitro. Furthermore, cross-linking studies reveal that Mge1-Hsp70 complex formation in mitochondria isolated from wild-type Mge1 cells is more susceptible to reactive oxygen species compared with mitochondria from Mge1-M155L cells. This novel oxidative sensor capability of yeast Mge1 might represent an evolutionarily conserved function, given that human recombinant dimeric Mge1 is also sensitive to H2O2.
Figures




References
-
- Azem A, Oppliger W, Lustig A, Jeno P, Feifel B, Schatz G, Horst M. The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J Biol Chem. 1997;272:20901–20906. - PubMed
-
- Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. - PubMed
-
- Cocheme HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

