Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle
- PMID: 23345615
- PMCID: PMC3597681
- DOI: 10.1093/nar/gkt015
Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle
Abstract
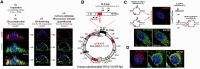
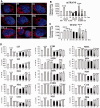
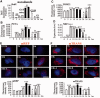
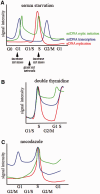
Nuclear (nDNA) and mitochondrial DNA (mtDNA) communication is essential for cell function, but it remains unclear whether the replication of these genomes is linked. We inspected human cells with a novel fluorescence in situ hybridization protocol (mitochondrial Transcription and Replication Imaging Protocol) that identifies mitochondrial structures engaged in initiation of mtDNA replication and unique transcript profiles, and reconstruct the temporal series of mitochondrial and nuclear events in single cells during the cell cycle. We show that mtDNA transcription and initiation of replication are prevalently coordinated with the cell cycle, preceding nuclear DNA synthesis, and being reactivated towards the end of S-phase. This coordination is achieved by modulating the fraction of mitochondrial structures that intiate mtDNA synthesis and/or contain transcript at a given time. Thus, although replication of the mitochondrial genome is active through the entire cell cycle, but in a limited fraction of mitochondrial structures, peaks of these activities are synchronized with nDNA synthesis. After release from blockage of mtDNA replication with either nocodazole or double thymidine treatment, prevalent mtDNA and nDNA synthesis occurred simultaneously, indicating that mitochondrial coordination with the nuclear phase can be adjusted in response to physiological alterations. These findings will help redefine other nuclear-mitochondrial links in cell function.
Figures



References
-
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. - PubMed
-
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. - PubMed
-
- Rotig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim. Biophys. Acta. 2009;1792:1103–1108. - PubMed
-
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

