Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity
- PMID: 23345616
- PMCID: PMC3597678
- DOI: 10.1093/nar/gkt012
Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity
Abstract
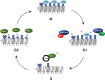
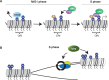
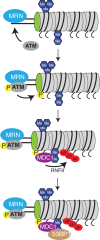
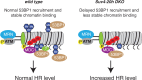
Maintenance of genomic integrity is essential to ensure normal organismal development and to prevent diseases such as cancer. Nuclear DNA is packaged into chromatin, and thus genome maintenance can be influenced by distinct chromatin environments. In particular, post-translational modifications of histones have emerged as key regulators of genomic integrity. Intense research during the past few years has revealed histone H4 lysine 20 methylation (H4K20me) as critically important for the biological processes that ensure genome integrity, such as DNA damage repair, DNA replication and chromatin compaction. The distinct H4K20 methylation states are mediated by SET8/PR-Set7 that catalyses monomethylation of H4K20, whereas SUV4-20H1 and SUV4-20H2 enzymes mediate further H4K20 methylation to H4K20me2 and H4K20me3. Disruption of these H4K20-specific histone methyltransferases leads to genomic instability, demonstrating the important functions of H4K20 methylation in genome maintenance. In this review, we explain molecular mechanisms underlying these defects and discuss novel ideas for furthering our understanding of genome maintenance in higher eukaryotes.
Figures
Similar articles
-
Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila.Nucleic Acids Res. 2016 Sep 6;44(15):7204-18. doi: 10.1093/nar/gkw333. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131378 Free PMC article.
-
The SUV4-20H Histone Methyltransferases in Health and Disease.Int J Mol Sci. 2022 Apr 25;23(9):4736. doi: 10.3390/ijms23094736. Int J Mol Sci. 2022. PMID: 35563127 Free PMC article. Review.
-
PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription.Genes Dev. 2012 Feb 15;26(4):325-37. doi: 10.1101/gad.177444.111. Genes Dev. 2012. PMID: 22345514 Free PMC article. Review.
-
Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks.PLoS One. 2012;7(11):e49211. doi: 10.1371/journal.pone.0049211. Epub 2012 Nov 28. PLoS One. 2012. PMID: 23209566 Free PMC article.
-
Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication.EMBO J. 2017 Sep 15;36(18):2726-2741. doi: 10.15252/embj.201796541. Epub 2017 Aug 4. EMBO J. 2017. PMID: 28778956 Free PMC article.
Cited by
-
Epigenetics and environment in breast cancer: New paradigms for anti-cancer therapies.Front Oncol. 2022 Sep 15;12:971288. doi: 10.3389/fonc.2022.971288. eCollection 2022. Front Oncol. 2022. PMID: 36185256 Free PMC article. Review.
-
The emerging role of lysine methyltransferase SETD8 in human diseases.Clin Epigenetics. 2016 Sep 22;8:102. doi: 10.1186/s13148-016-0268-4. eCollection 2016. Clin Epigenetics. 2016. PMID: 27688818 Free PMC article. Review.
-
Histone methylation and the DNA damage response.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:37-47. doi: 10.1016/j.mrrev.2017.09.003. Epub 2017 Sep 23. Mutat Res Rev Mutat Res. 2019. PMID: 31395347 Free PMC article. Review.
-
The association between rs16917496 T/C polymorphism of SET8 gene and cancer risk in Asian populations: a meta-analysis.Biosci Rep. 2018 Nov 13;38(6):BSR20180702. doi: 10.1042/BSR20180702. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30341251 Free PMC article.
-
Short tandem repeats are important contributors to silencer elements in T cells.Nucleic Acids Res. 2023 Jun 9;51(10):4845-4866. doi: 10.1093/nar/gkad187. Nucleic Acids Res. 2023. PMID: 36929452 Free PMC article.
References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. - PubMed
-
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

