Same, same but different: symbiotic bacterial associations in GBR sponges
- PMID: 23346080
- PMCID: PMC3548243
- DOI: 10.3389/fmicb.2012.00444
Same, same but different: symbiotic bacterial associations in GBR sponges
Abstract
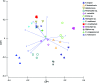
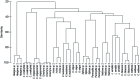
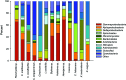
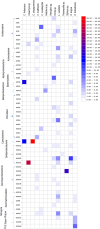
Symbioses in marine sponges involve diverse consortia of microorganisms that contribute to the health and ecology of their hosts. The microbial communities of 13 taxonomically diverse Great Barrier Reef (GBR) sponge species were assessed by DGGE and 16S rRNA gene sequencing to determine intra and inter species variation in bacterial symbiont composition. Microbial profiling revealed communities that were largely conserved within different individuals of each species with intra species similarity ranging from 65-100%. 16S rRNA gene sequencing revealed that the communities were dominated by Proteobacteria, Chloroflexi, Acidobacteria, Actinobacteria, Nitrospira, and Cyanobacteria. Sponge-associated microbes were also highly host-specific with no operational taxonomic units (OTUs) common to all species and the most ubiquitous OTU found in only 5 of the 13 sponge species. In total, 91% of the OTUs were restricted to a single sponge species. However, GBR sponge microbes were more closely related to other sponge-derived bacteria than they were to environmental communities with sequences falling within 50 of the 173 previously defined sponge-(or sponge-coral) specific sequence clusters (SC). These SC spanned the Acidobacteria, Actinobacteria, Proteobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Nitrospira, and the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. The number of sequences assigned to these sponge-specific clusters across all species ranged from 0 to 92%. No relationship between host phylogeny and symbiont communities were observed across the different sponge orders, although the highest level of similarity was detected in two closely related Xestospongia species. This study identifies the core microbial inhabitants in a range of GBR sponges thereby providing the basis for future studies on sponge symbiotic function and research aiming to predict how sponge holobionts will respond to environmental perturbation.
Keywords: Great Barrier Reef; diversity; microorganism; sponge; symbiont.
Figures

Similar articles
-
Symbiotic prokaryotic communities from different populations of the giant barrel sponge, Xestospongia muta.Microbiologyopen. 2013 Dec;2(6):938-52. doi: 10.1002/mbo3.135. Epub 2013 Sep 30. Microbiologyopen. 2013. PMID: 24124112 Free PMC article.
-
Phylogenetic diversity and community structure of the symbionts associated with the coralline sponge Astrosclera willeyana of the Great Barrier Reef.Microb Ecol. 2013 Apr;65(3):740-52. doi: 10.1007/s00248-013-0212-5. Epub 2013 Mar 24. Microb Ecol. 2013. PMID: 23525793
-
Microbial diversity in the coralline sponge Vaceletia crypta.Antonie Van Leeuwenhoek. 2013 May;103(5):1041-56. doi: 10.1007/s10482-013-9884-6. Epub 2013 Jan 30. Antonie Van Leeuwenhoek. 2013. PMID: 23361993
-
Diversity and biotechnological potential of the sponge-associated microbial consortia.J Ind Microbiol Biotechnol. 2006 Jul;33(7):545-51. doi: 10.1007/s10295-006-0123-2. Epub 2006 Apr 22. J Ind Microbiol Biotechnol. 2006. PMID: 16761166 Review.
-
Fungal association with sessile marine invertebrates.Front Microbiol. 2014 May 15;5:228. doi: 10.3389/fmicb.2014.00228. eCollection 2014. Front Microbiol. 2014. PMID: 24860565 Free PMC article. Review.
Cited by
-
A novel uncultured heterotrophic bacterial associate of the cyanobacterium Moorea producens JHB.BMC Microbiol. 2016 Aug 30;16(1):198. doi: 10.1186/s12866-016-0817-1. BMC Microbiol. 2016. PMID: 27577966 Free PMC article.
-
Pyrosequencing reveals sponge specific bacterial communities in marine sponges of Red Sea, Saudi Arabia.Saudi J Biol Sci. 2020 Jan;27(1):67-73. doi: 10.1016/j.sjbs.2019.05.002. Epub 2019 May 7. Saudi J Biol Sci. 2020. PMID: 31889819 Free PMC article.
-
Mini review: antimicrobial compounds produced by bacteria associated with marine invertebrates.Folia Microbiol (Praha). 2025 Apr;70(2):271-292. doi: 10.1007/s12223-024-01209-5. Epub 2024 Oct 24. Folia Microbiol (Praha). 2025. PMID: 39446239 Review.
-
Pyrosequencing revealed shifts of prokaryotic communities between healthy and disease-like tissues of the Red Sea sponge Crella cyathophora.PeerJ. 2015 Jun 11;3:e890. doi: 10.7717/peerj.890. eCollection 2015. PeerJ. 2015. PMID: 26082867 Free PMC article.
-
Two distinct microbial communities revealed in the sponge Cinachyrella.Front Microbiol. 2014 Nov 4;5:581. doi: 10.3389/fmicb.2014.00581. eCollection 2014. Front Microbiol. 2014. PMID: 25408689 Free PMC article.
References
-
- Anderson M. J., Gorley R. N., Clarke K. R. (2008). PERMANOVA+ for PRIMER: Guide to software and statistical methods, in PRIMER-E, (Plymouth: ).
-
- Enticknap J. J., Kelly M., Peraud O., Hill R. T. (2006). Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72, 3724–3732 10.1128/AEM.72.5.3724-3732.2006 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

