Identification of viral pathogen diversity in sewage sludge by metagenome analysis
- PMID: 23346855
- PMCID: PMC3963146
- DOI: 10.1021/es305181x
Identification of viral pathogen diversity in sewage sludge by metagenome analysis
Abstract
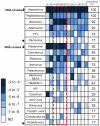
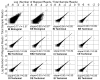
The large diversity of viruses that exist in human populations are potentially excreted into sewage collection systems and concentrated in sewage sludge. In the U.S., the primary fate of processed sewage sludge (class B biosolids) is application to agricultural land as a soil amendment. To characterize and understand infectious risks associated with land application, and to describe the diversity of viruses in human populations, shotgun viral metagenomics was applied to 10 sewage sludge samples from 5 wastewater treatment plants throughout the continental U.S, each serving between 100,000 and 1,000,000 people. Nearly 330 million DNA sequences were produced and assembled, and annotation resulted in identifying 43 (26 DNA, 17 RNA) different types of human viruses in sewage sludge. Novel insights include the high abundance of newly emerging viruses (e.g., Coronavirus HKU1, Klassevirus, and Cosavirus) the strong representation of respiratory viruses, and the relatively minor abundance and occurrence of Enteroviruses. Viral metagenome sequence annotations were reproducible and independent PCR-based identification of selected viruses suggests that viral metagenomes were a conservative estimate of the true viral occurrence and diversity. These results represent the most complete description of human virus diversity in any wastewater sample to date, provide engineers and environmental scientists with critical information on important viral agents and routes of infection from exposure to wastewater and sewage sludge, and represent a significant leap forward in understanding the pathogen content of class B biosolids.
Figures


References
-
- USEPA . Ambient Water Quality Criteria for Bacteria. 1986. EPA440/5-84-002.
-
- Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environmental Microbiology. 2009;11:2806–2820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

