Characterization of the basal angiosperm Aristolochia fimbriata: a potential experimental system for genetic studies
- PMID: 23347749
- PMCID: PMC3621149
- DOI: 10.1186/1471-2229-13-13
Characterization of the basal angiosperm Aristolochia fimbriata: a potential experimental system for genetic studies
Abstract
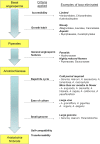
Background: Previous studies in basal angiosperms have provided insight into the diversity within the angiosperm lineage and helped to polarize analyses of flowering plant evolution. However, there is still not an experimental system for genetic studies among basal angiosperms to facilitate comparative studies and functional investigation. It would be desirable to identify a basal angiosperm experimental system that possesses many of the features found in existing plant model systems (e.g., Arabidopsis and Oryza).
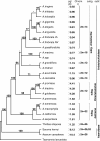
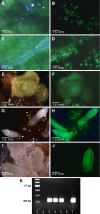
Results: We have considered all basal angiosperm families for general characteristics important for experimental systems, including availability to the scientific community, growth habit, and membership in a large basal angiosperm group that displays a wide spectrum of phenotypic diversity. Most basal angiosperms are woody or aquatic, thus are not well-suited for large scale cultivation, and were excluded. We further investigated members of Aristolochiaceae for ease of culture, life cycle, genome size, and chromosome number. We demonstrated self-compatibility for Aristolochia elegans and A. fimbriata, and transformation with a GFP reporter construct for Saruma henryi and A. fimbriata. Furthermore, A. fimbriata was easily cultivated with a life cycle of just three months, could be regenerated in a tissue culture system, and had one of the smallest genomes among basal angiosperms. An extensive multi-tissue EST dataset was produced for A. fimbriata that includes over 3.8 million 454 sequence reads.
Conclusions: Aristolochia fimbriata has numerous features that facilitate genetic studies and is suggested as a potential model system for use with a wide variety of technologies. Emerging genetic and genomic tools for A. fimbriata and closely related species can aid the investigation of floral biology, developmental genetics, biochemical pathways important in plant-insect interactions as well as human health, and various other features present in early angiosperms.
Figures





References
-
- Kellogg EA, Shaffer HB. Model organisms in evolutionary studies. Syst Biol. 1993;42(4):409–414. doi: 10.1093/sysbio/42.4.409. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

