Phylogenetic and ecologic perspectives of a monkeypox outbreak, southern Sudan, 2005
- PMID: 23347770
- PMCID: PMC3559062
- DOI: 10.3201/eid1902.121220
Phylogenetic and ecologic perspectives of a monkeypox outbreak, southern Sudan, 2005
Abstract
Identification of human monkeypox cases during 2005 in southern Sudan (now South Sudan) raised several questions about the natural history of monkeypox virus (MPXV) in Africa. The outbreak area, characterized by seasonally dry riverine grasslands, is not identified as environmentally suitable for MPXV transmission. We examined possible origins of this outbreak by performing phylogenetic analysis of genome sequences of MPXV isolates from the outbreak in Sudan and from differing localities. We also compared the environmental suitability of study localities for monkeypox transmission. Phylogenetically, the viruses isolated from Sudan outbreak specimens belong to a clade identified in the Congo Basin. This finding, added to the political instability of the area during the time of the outbreak, supports the hypothesis of importation by infected animals or humans entering Sudan from the Congo Basin, and person-to-person transmission of virus, rather than transmission of indigenous virus from infected animals to humans.
Figures

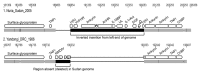
References
-
- von Magnus P, Andersen EK, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathol Microbiol Scand. 1959;46:156–76. 10.1111/j.1699-0463.1959.tb00328.x - DOI
-
- Ježek Z, Fenner F. Human monkeypox. New York: Karger; 1988.
-
- Reynolds MG, Carroll DS, Olson VA, Hughes C, Galley J, Likos AM, et al. A silent enzootic of an orthopoxvirus in Ghana, West Africa: evidence for multi-species involvement in the absence of widespread human disease. Am J Trop Med Hyg. 2010;82:746–54 and. 10.4269/ajtmh.2010.09-0716 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
