Incorporation of a viral DNA-packaging motor channel in lipid bilayers for real-time, single-molecule sensing of chemicals and double-stranded DNA
- PMID: 23348364
- PMCID: PMC3866906
- DOI: 10.1038/nprot.2013.001
Incorporation of a viral DNA-packaging motor channel in lipid bilayers for real-time, single-molecule sensing of chemicals and double-stranded DNA
Abstract
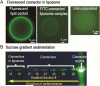
Over the past decade, nanopores have rapidly emerged as stochastic biosensors. This protocol describes the cloning, expression and purification of the channel of the bacteriophage phi29 DNA-packaging nanomotor and its subsequent incorporation into lipid membranes for single-pore sensing of double-stranded DNA (dsDNA) and chemicals. The membrane-embedded phi29 nanochannel remains functional and structurally intact under a range of conditions. When ions and macromolecules translocate through this nanochannel, reliable fingerprint changes in conductance are observed. Compared with other well-studied biological pores, the phi29 nanochannel has a larger cross-sectional area, which enables the translocation of dsDNA. Furthermore, specific amino acids can be introduced by site-directed mutagenesis within the large cavity of the channel to conjugate receptors that are able to bind specific ligands or analytes for desired applications. The lipid membrane-embedded nanochannel system has immense potential nanotechnological and biomedical applications in bioreactors, environmental sensing, drug monitoring, controlled drug delivery, early disease diagnosis and high-throughput DNA sequencing. The total time required for completing one round of this protocol is around 1 month.
Figures
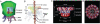




Similar articles
-
Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids.ACS Nano. 2013 Apr 23;7(4):3315-23. doi: 10.1021/nn400020z. Epub 2013 Mar 25. ACS Nano. 2013. PMID: 23488809 Free PMC article.
-
Real-time sensing and discrimination of single chemicals using the channel of phi29 DNA packaging nanomotor.ACS Nano. 2012 Apr 24;6(4):3251-61. doi: 10.1021/nn3001615. Epub 2012 Apr 9. ACS Nano. 2012. PMID: 22458779 Free PMC article.
-
Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores.Nat Nanotechnol. 2009 Nov;4(11):765-72. doi: 10.1038/nnano.2009.259. Epub 2009 Sep 27. Nat Nanotechnol. 2009. PMID: 19893523 Free PMC article.
-
Ultrastable pRNA hexameric ring gearing hexameric phi29 DNA-packaging motor by revolving without rotating and coiling.Curr Opin Biotechnol. 2013 Aug;24(4):581-90. doi: 10.1016/j.copbio.2013.03.019. Epub 2013 May 14. Curr Opin Biotechnol. 2013. PMID: 23683853 Free PMC article. Review.
-
Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy.Ann Biomed Eng. 2009 Oct;37(10):2064-81. doi: 10.1007/s10439-009-9723-0. Epub 2009 Jun 4. Ann Biomed Eng. 2009. PMID: 19495981 Free PMC article. Review.
Cited by
-
Structure and Dynamics of Thermosensitive pDNA Polyplexes Studied by Time-Resolved Fluorescence Spectroscopy.Biomacromolecules. 2020 Jan 13;21(1):73-88. doi: 10.1021/acs.biomac.9b00896. Epub 2019 Sep 24. Biomacromolecules. 2020. PMID: 31500418 Free PMC article.
-
Construction of an aerolysin nanopore in a lipid bilayer for single-oligonucleotide analysis.Nat Protoc. 2017 Sep;12(9):1901-1911. doi: 10.1038/nprot.2017.077. Epub 2017 Aug 24. Nat Protoc. 2017. PMID: 28837133
-
Nanobiomotors of archaeal DNA repair machineries: current research status and application potential.Cell Biosci. 2014 Jun 25;4:32. doi: 10.1186/2045-3701-4-32. eCollection 2014. Cell Biosci. 2014. PMID: 24995126 Free PMC article. Review.
-
Construction of RNA nanotubes.Nano Res. 2019 Aug;12(8):1952-1958. doi: 10.1007/s12274-019-2463-z. Epub 2019 Jul 11. Nano Res. 2019. PMID: 32153728 Free PMC article.
-
Cell-free production of a functional oligomeric form of a Chlamydia major outer-membrane protein (MOMP) for vaccine development.J Biol Chem. 2017 Sep 8;292(36):15121-15132. doi: 10.1074/jbc.M117.784561. Epub 2017 Jul 24. J Biol Chem. 2017. PMID: 28739800 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

