ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1
- PMID: 23348497
- PMCID: PMC3746320
- DOI: 10.1016/j.cbi.2012.12.018
ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1
Abstract
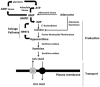
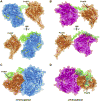
Gout, a common form of inflammatory arthritis, is strongly associated with elevated uric acid concentrations in the blood (hyperuricemia). A recent study in Icelanders identified a rare missense single nucleotide polymorphism (SNP) in the ALDH16A1 gene, ALDH16A1*2, to be associated with gout and serum uric acid levels. ALDH16A1 is a novel and rather unique member of the ALDH superfamily in relation to its gene and protein structures. ALDH16 genes are present in fish, amphibians, protista, bacteria but absent from archaea, fungi and plants. In most mammalian species, two ALDH16A1 spliced variants (ALDH16A1, long form and ALDH16A1_v2, short form) have been identified and both are expressed in HepG-2, HK-2 and HK-293 human cell lines. The ALDH16 proteins contain two ALDH domains (as opposed to one in the other members of the superfamily), four transmembrane and one coiled-coil domains. The active site of ALDH16 proteins from bacterial, frog and lower animals contain the catalytically important cysteine residue (Cys-302); this residue is absent from the mammalian and fish orthologs. Molecular modeling predicts that both the short and long forms of human ALDH16A1 protein would lack catalytic activity but may interact with the hypoxanthine-guanine phosphoribosyltransferase (HPRT1) protein, a key enzyme involved in uric acid metabolism and gout. Interestingly, such protein-protein interactions with HPRT1 are predicted to be impaired for the long or short forms of ALDH16A1*2. These results lead to the intriguing possibility that association between ALDH16A1 and HPRT1 may be required for optimal HPRT activity with disruption of this interaction possibly contributing to the hyperuricemia seen in ALDH16A1*2 carriers.
Copyright © 2013 Elsevier Ireland Ltd. All rights reserved.
Conflict of interest statement
Figures




Similar articles
-
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.J Mol Biol. 2019 Feb 1;431(3):524-541. doi: 10.1016/j.jmb.2018.11.030. Epub 2018 Dec 7. J Mol Biol. 2019. PMID: 30529746 Free PMC article.
-
Expression, purification and crystallization of the novel Xenopus tropicalis ALDH16B1, a homologue of human ALDH16A1.Chem Biol Interact. 2019 May 1;304:168-172. doi: 10.1016/j.cbi.2019.03.009. Epub 2019 Mar 17. Chem Biol Interact. 2019. PMID: 30894314 Free PMC article.
-
Transcriptomic analysis and plasma metabolomics in Aldh16a1-null mice reveals a potential role of ALDH16A1 in renal function.Chem Biol Interact. 2017 Oct 1;276:15-22. doi: 10.1016/j.cbi.2017.02.013. Epub 2017 Feb 28. Chem Biol Interact. 2017. PMID: 28254523 Free PMC article.
-
Recent insights into the pathogenesis of hyperuricaemia and gout.Hum Mol Genet. 2009 Oct 15;18(R2):R177-84. doi: 10.1093/hmg/ddp369. Hum Mol Genet. 2009. PMID: 19808794 Review.
-
[Deficiencies of hypoxanthine guanine phosphoribosyltransferase (HPRT)].Nihon Rinsho. 2008 Apr;66(4):687-93. Nihon Rinsho. 2008. PMID: 18409516 Review. Japanese.
Cited by
-
Comparison Between Early-Onset and Common Gout: A Systematic Literature Review.Rheumatol Ther. 2023 Aug;10(4):809-823. doi: 10.1007/s40744-023-00565-x. Epub 2023 Jun 19. Rheumatol Ther. 2023. PMID: 37335432 Free PMC article. Review.
-
ALDH3A1 Plays a Functional Role in Maintenance of Corneal Epithelial Homeostasis.PLoS One. 2016 Jan 11;11(1):e0146433. doi: 10.1371/journal.pone.0146433. eCollection 2016. PLoS One. 2016. PMID: 26751691 Free PMC article.
-
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.J Biol Chem. 2016 Nov 11;291(46):24065-24075. doi: 10.1074/jbc.M116.756965. Epub 2016 Sep 27. J Biol Chem. 2016. PMID: 27679491 Free PMC article.
-
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.J Mol Biol. 2019 Feb 1;431(3):524-541. doi: 10.1016/j.jmb.2018.11.030. Epub 2018 Dec 7. J Mol Biol. 2019. PMID: 30529746 Free PMC article.
-
Expression, purification and crystallization of the novel Xenopus tropicalis ALDH16B1, a homologue of human ALDH16A1.Chem Biol Interact. 2019 May 1;304:168-172. doi: 10.1016/j.cbi.2019.03.009. Epub 2019 Mar 17. Chem Biol Interact. 2019. PMID: 30894314 Free PMC article.
References
-
- Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13:489–498. - PubMed
-
- Brew K, Vanaman TC, Hill RL. Comparison of the amino acid sequence of bovine alpha-lactalbumin and hens egg white lysozyme. J Biol Chem. 1967;242:3747–3749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

