Genome-wide analysis reveals selection for important traits in domestic horse breeds
- PMID: 23349635
- PMCID: PMC3547851
- DOI: 10.1371/journal.pgen.1003211
Genome-wide analysis reveals selection for important traits in domestic horse breeds
Abstract
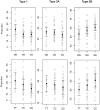
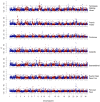
Intense selective pressures applied over short evolutionary time have resulted in homogeneity within, but substantial variation among, horse breeds. Utilizing this population structure, 744 individuals from 33 breeds, and a 54,000 SNP genotyping array, breed-specific targets of selection were identified using an F(ST)-based statistic calculated in 500-kb windows across the genome. A 5.5-Mb region of ECA18, in which the myostatin (MSTN) gene was centered, contained the highest signature of selection in both the Paint and Quarter Horse. Gene sequencing and histological analysis of gluteal muscle biopsies showed a promoter variant and intronic SNP of MSTN were each significantly associated with higher Type 2B and lower Type 1 muscle fiber proportions in the Quarter Horse, demonstrating a functional consequence of selection at this locus. Signatures of selection on ECA23 in all gaited breeds in the sample led to the identification of a shared, 186-kb haplotype including two doublesex related mab transcription factor genes (DMRT2 and 3). The recent identification of a DMRT3 mutation within this haplotype, which appears necessary for the ability to perform alternative gaits, provides further evidence for selection at this locus. Finally, putative loci for the determination of size were identified in the draft breeds and the Miniature horse on ECA11, as well as when signatures of selection surrounding candidate genes at other loci were examined. This work provides further evidence of the importance of MSTN in racing breeds, provides strong evidence for selection upon gait and size, and illustrates the potential for population-based techniques to find genomic regions driving important phenotypes in the modern horse.
Conflict of interest statement
Equinome Ltd. (EWH, Director) has been granted a license for commercial use of MSTN data as contained within patent applications: U.S. Provisional Serial Number 61/136553; Irish Patent Application Numbers 2008/0735 and 2010/0151; and Patent Cooperation Treaty number PCT/IE2009/000062. The PCT publication WO2010/029527A published 18 March 2010. Title: “A method for predicting athletics performance potential” and U.S. publication US2011/0262915 published 27 October 2011. Title: “Method for predicting the athletic performance potential of a subject.” EWH, NO, and BAM are named on the applications. MMB works for The Genetic Edge, previously published a paper on the association between SNPs in the MSTN region and best racing distance for elite Thoroughbred horses [46], and uses these markers in commercial tests. LSA and GL are co-applicants on a patent application concerning the commercial utilization of the DMRT3 mutation. These commercial ventures had no influence on the interpretation of the results relating to myostatin or gait presented in the paper.
Figures









References
-
- Outram AK, Stear NA, Bendrey R, Olsen S, Kasparov A, et al. (2009) The earliest horse harnessing and milking. Science 323: 1332–1335. - PubMed
-
- Olsson M, Meadows JR, Truve K, Rosengren Pielberg G, Puppo F, et al. (2011) A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet 7: e1001332 doi:10.1371/journal.pgen.1001332. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

