Cloning, annotation and developmental expression of the chicken intestinal MUC2 gene
- PMID: 23349743
- PMCID: PMC3549977
- DOI: 10.1371/journal.pone.0053781
Cloning, annotation and developmental expression of the chicken intestinal MUC2 gene
Abstract
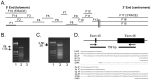
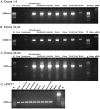
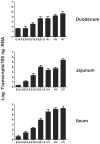
Intestinal mucin 2 (MUC2) encodes a heavily glycosylated, gel-forming mucin, which creates an important protective mucosal layer along the gastrointestinal tract in humans and other species. This first line of defense guards against attacks from microorganisms and is integral to the innate immune system. As a first step towards characterizing the innate immune response of MUC2 in different species, we report the cloning of a full-length, 11,359 bp chicken MUC2 cDNA, and describe the genomic organization and functional annotation of this complex, 74.5 kb locus. MUC2 contains 64 exons and demonstrates distinct spatiotemporal expression profiles throughout development in the gastrointestinal tract; expression increases with gestational age and from anterior to posterior along the gut. The chicken protein has a similar domain organization as the human orthologue, with a signal peptide and several von Willebrand domains in the N-terminus and the characteristic cystine knot at the C-terminus. The PTS domain of the chicken MUC2 protein spans ∼1600 amino acids and is interspersed with four CysD motifs. However, the PTS domain in the chicken diverges significantly from the human orthologue; although the chicken domain is shorter, the repetitive unit is 69 amino acids in length, which is three times longer than the human. The amino acid composition shows very little similarity to the human motif, which potentially contributes to differences in the innate immune response between species, as glycosylation across this rapidly evolving domain provides much of the musical barrier. Future studies of the function of MUC2 in the innate immune response system in chicken could provide an important model organism to increase our understanding of the biological significance of MUC2 in host defense and highlight the potential of the chicken for creating new immune-based therapies.
Conflict of interest statement
Figures





Similar articles
-
An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins.BMC Genomics. 2006 Aug 3;7:197. doi: 10.1186/1471-2164-7-197. BMC Genomics. 2006. PMID: 16887038 Free PMC article.
-
Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor.J Biol Chem. 1994 Jan 28;269(4):2440-6. J Biol Chem. 1994. PMID: 8300571
-
Gastrointestinal expression and partial cDNA cloning of murine Muc2.Am J Physiol. 1999 Jan;276(1):G115-24. doi: 10.1152/ajpgi.1999.276.1.G115. Am J Physiol. 1999. PMID: 9886986
-
The MUC2 gene product: a human intestinal mucin.Int J Biochem Cell Biol. 1998 Jul;30(7):797-801. doi: 10.1016/s1357-2725(98)00028-4. Int J Biochem Cell Biol. 1998. PMID: 9722984 Review.
-
Mucin 2 (MUC2) promoter characterization: an overview.Cell Tissue Res. 2018 Dec;374(3):455-463. doi: 10.1007/s00441-018-2916-9. Epub 2018 Sep 14. Cell Tissue Res. 2018. PMID: 30218241 Review.
Cited by
-
Effect of phytase on nutrient digestibility and expression of intestinal tight junction and nutrient transporter genes in pigs.J Anim Sci. 2020 Jul 1;98(7):skaa206. doi: 10.1093/jas/skaa206. J Anim Sci. 2020. PMID: 32607561 Free PMC article.
-
Identification of Potential Biomarkers for Gut Barrier Failure in Broiler Chickens.Front Vet Sci. 2015 May 26;2:14. doi: 10.3389/fvets.2015.00014. eCollection 2015. Front Vet Sci. 2015. PMID: 26664943 Free PMC article.
-
Coccidia-Microbiota Interactions and Their Effects on the Host.Front Cell Infect Microbiol. 2021 Oct 1;11:751481. doi: 10.3389/fcimb.2021.751481. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34660347 Free PMC article. Review.
-
Changes with age in density of goblet cells in the small intestine of broiler chicks.Poult Sci. 2020 May;99(5):2342-2348. doi: 10.1016/j.psj.2019.12.052. Epub 2020 Mar 18. Poult Sci. 2020. PMID: 32359569 Free PMC article.
-
Butyric and valeric glycerides alleviate sub-clinical necrotic enteritis effect on performance and gut health of broiler chickens.Poult Sci. 2025 Jun 13;104(9):105441. doi: 10.1016/j.psj.2025.105441. Online ahead of print. Poult Sci. 2025. PMID: 40577949 Free PMC article.
References
-
- Hollingsworth MA, Swanson BJ (2004) Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4: 45–60. - PubMed
-
- McGuckin MA, Linden SK, Sutton P, Florin TH (2011) Mucin dynamics and enteric pathogens. Nat Rev Microbiology 9: 265–278. - PubMed
-
- Larsson JM, Karlsson H, Sjovall H, Hansson GC (2009) A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19: 756–766. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

