De novo transcriptome sequencing and analysis for Venturia inaequalis, the devastating apple scab pathogen
- PMID: 23349770
- PMCID: PMC3547962
- DOI: 10.1371/journal.pone.0053937
De novo transcriptome sequencing and analysis for Venturia inaequalis, the devastating apple scab pathogen
Abstract
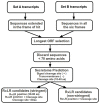
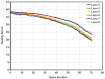
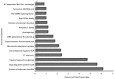
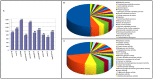
Venturia inaequalis is the causal agent of apple scab, one of the most devastating diseases of apple. Due to several distinct features, it has emerged as a model fungal pathogen to study various aspects of hemibiotrophic plant pathogen interactions. The present study reports de novo assembling, annotation and characterization of the transcriptome of V. inaequalis. Venturia transcripts expressed during its growth on laboratory medium and that expressed during its biotrophic stage of infection on apple were sequenced using Illumina RNAseq technology. A total of 94,350,055 reads (50 bp read length) specific to Venturia were obtained after filtering. The reads were assembled into 62,061 contigs representing 24,571 unique genes. GO analysis suggested prevalence of genes associated with biological process categories like metabolism, transport and response to stimulus. Genes associated with molecular function like binding, catalytic activities and transferase activities were found in majority. EC and KEGG pathway analyses suggested prevalence of genes encoding kinases, proteases, glycoside hydrolases, cutinases, cytochrome P450 and transcription factors. The study has identified several putative pathogenicity determinants and candidate effectors in V. inaequalis. A large number of transcripts encoding membrane transporters were identified and comparative analysis revealed that the number of transporters encoded by Venturia is significantly more as compared to that encoded by several other important plant fungal pathogens. Phylogenomics analysis indicated that V. inaequalis is closely related to Pyrenophora tritici-repentis (the causal organism of tan spot of wheat). In conclusion, the findings from this study provide a better understanding of the biology of the apple scab pathogen and have identified candidate genes/functions required for its pathogenesis. This work lays the foundation for facilitating further research towards understanding this host-pathogen interaction.
Conflict of interest statement
Figures





Similar articles
-
The Venturia inaequalis effector repertoire is dominated by expanded families with predicted structural similarity, but unrelated sequence, to avirulence proteins from other plant-pathogenic fungi.BMC Biol. 2022 Nov 3;20(1):246. doi: 10.1186/s12915-022-01442-9. BMC Biol. 2022. PMID: 36329441 Free PMC article.
-
Comparative analysis of the predicted secretomes of Rosaceae scab pathogens Venturia inaequalis and V. pirina reveals expanded effector families and putative determinants of host range.BMC Genomics. 2017 May 2;18(1):339. doi: 10.1186/s12864-017-3699-1. BMC Genomics. 2017. PMID: 28464870 Free PMC article.
-
Venturia inaequalis: the causal agent of apple scab.Mol Plant Pathol. 2011 Feb;12(2):105-22. doi: 10.1111/j.1364-3703.2010.00656.x. Epub 2010 Aug 26. Mol Plant Pathol. 2011. PMID: 21199562 Free PMC article.
-
The Venturia apple pathosystem: pathogenicity mechanisms and plant defense responses.J Biomed Biotechnol. 2009;2009:680160. doi: 10.1155/2009/680160. Epub 2010 Jan 28. J Biomed Biotechnol. 2009. PMID: 20150969 Free PMC article. Review.
-
Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus.Annu Rev Phytopathol. 2011;49:391-413. doi: 10.1146/annurev-phyto-072910-095339. Annu Rev Phytopathol. 2011. PMID: 21599495 Review.
Cited by
-
Phenotyping, genetics, and "-omics" approaches to unravel and introgress enhanced resistance against apple scab (Venturia inaequalis) in apple cultivars (Malus × domestica).Hortic Res. 2024 Jan 10;11(2):uhae002. doi: 10.1093/hr/uhae002. eCollection 2024 Feb. Hortic Res. 2024. PMID: 38371632 Free PMC article.
-
Dual RNA-seq reveals Meloidogyne graminicola transcriptome and candidate effectors during the interaction with rice plants.Mol Plant Pathol. 2016 Aug;17(6):860-74. doi: 10.1111/mpp.12334. Epub 2016 Apr 4. Mol Plant Pathol. 2016. PMID: 26610268 Free PMC article.
-
Augmentation of crop productivity through interventions of omics technologies in India: challenges and opportunities.3 Biotech. 2018 Nov;8(11):454. doi: 10.1007/s13205-018-1473-y. Epub 2018 Oct 19. 3 Biotech. 2018. PMID: 30370195 Free PMC article. Review.
-
The Venturia inaequalis effector repertoire is dominated by expanded families with predicted structural similarity, but unrelated sequence, to avirulence proteins from other plant-pathogenic fungi.BMC Biol. 2022 Nov 3;20(1):246. doi: 10.1186/s12915-022-01442-9. BMC Biol. 2022. PMID: 36329441 Free PMC article.
-
Computational Analysis of HTS Data and Its Application in Plant Pathology.Methods Mol Biol. 2022;2536:275-307. doi: 10.1007/978-1-0716-2517-0_17. Methods Mol Biol. 2022. PMID: 35819611
References
-
- Jha G, Thakur K, Thakur P (2009) The Venturia apple pathosystem: pathogenicity mechanisms and plant defense responses. J Biomed Biotech doi:10.1155/2009/680160. - DOI - PMC - PubMed
-
- MacHardy WE (1996) Apple Scab, Biology, Epidemiology, and Management. St. Paul, MinnUSA: APS Press.
-
- Gupta GK (1985) Recent trends in forecasting and control of apple scab (Venturia inaequalis). Pesticides 19: 19–31.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

