Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory
- PMID: 23349810
- PMCID: PMC3549990
- DOI: 10.1371/journal.pone.0054144
Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory
Abstract
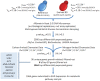
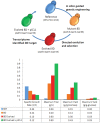
Saccharomyces cerevisiae is the most well characterized eukaryote, the preferred microbial cell factory for the largest industrial biotechnology product (bioethanol), and a robust commerically compatible scaffold to be exploitted for diverse chemical production. Succinic acid is a highly sought after added-value chemical for which there is no native pre-disposition for production and accmulation in S. cerevisiae. The genome-scale metabolic network reconstruction of S. cerevisiae enabled in silico gene deletion predictions using an evolutionary programming method to couple biomass and succinate production. Glycine and serine, both essential amino acids required for biomass formation, are formed from both glycolytic and TCA cycle intermediates. Succinate formation results from the isocitrate lyase catalyzed conversion of isocitrate, and from the α-keto-glutarate dehydrogenase catalyzed conversion of α-keto-glutarate. Succinate is subsequently depleted by the succinate dehydrogenase complex. The metabolic engineering strategy identified included deletion of the primary succinate consuming reaction, Sdh3p, and interruption of glycolysis derived serine by deletion of 3-phosphoglycerate dehydrogenase, Ser3p/Ser33p. Pursuing these targets, a multi-gene deletion strain was constructed, and directed evolution with selection used to identify a succinate producing mutant. Physiological characterization coupled with integrated data analysis of transcriptome data in the metabolically engineered strain were used to identify 2(nd)-round metabolic engineering targets. The resulting strain represents a 30-fold improvement in succinate titer, and a 43-fold improvement in succinate yield on biomass, with only a 2.8-fold decrease in the specific growth rate compared to the reference strain. Intuitive genetic targets for either over-expression or interruption of succinate producing or consuming pathways, respectively, do not lead to increased succinate. Rather, we demonstrate how systems biology tools coupled with directed evolution and selection allows non-intuitive, rapid and substantial re-direction of carbon fluxes in S. cerevisiae, and hence show proof of concept that this is a potentially attractive cell factory for over-producing different platform chemicals.
Conflict of interest statement
Figures


References
-
- Otero JM, Panagiotou G, Olsson L (2007) Fueling Industrial Biotechnology Through Bioethanol. Adv Biochem Eng Biotechnol 108: 1–40. - PubMed
-
- Nielsen J (2001) Metabolic engineering. Appl Microbiol Biotechnol 55(3): 263–283. - PubMed
-
- Covert MW, Schilling CH, Famili I, Edwards JS, Goryanin II, et al. (2001) Metabolic modeling of microbial strains in silico. Trends Biochem Sci 26(3): 179–186. - PubMed
-
- Nielsen J, Jewett MC (2008) Impact of systems biology on metabolic engineering of Saccharomyces cerevisiae . FEMS Yeast Res 8(1): 122–131. - PubMed
-
- Bro C, Regenberg B, Förster J, Nielsen J (2005) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metabolic Engineering 8(2): 102–111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

