Finding protein-coding genes through human polymorphisms
- PMID: 23349826
- PMCID: PMC3551959
- DOI: 10.1371/journal.pone.0054210
Finding protein-coding genes through human polymorphisms
Abstract
Human gene catalogs are fundamental to the study of human biology and medicine. But they are all based on open reading frames (ORFs) in a reference genome sequence (with allowance for introns). Individual genomes, however, are polymorphic: their sequences are not identical. There has been much research on how polymorphism affects previously-identified genes, but no research has been done on how it affects gene identification itself. We computationally predict protein-coding genes in a straightforward manner, by finding long ORFs in mRNA sequences aligned to the reference genome. We systematically test the effect of known polymorphisms with this procedure. Polymorphisms can not only disrupt ORFs, they can also create long ORFs that do not exist in the reference sequence. We found 5,737 putative protein-coding genes that do not exist in the reference, whose protein-coding status is supported by homology to known proteins. On average 10% of these genes are located in the genomic regions devoid of annotated genes in 12 other catalogs. Our statistical analysis showed that these ORFs are unlikely to occur by chance.
Conflict of interest statement
Figures
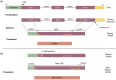
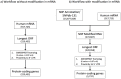



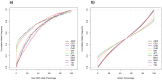
 -axis) in panel (b) is based on Allele1, chosen arbitrarily.
-axis) in panel (b) is based on Allele1, chosen arbitrarily.Similar articles
-
Distinguishing protein-coding and noncoding genes in the human genome.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19428-33. doi: 10.1073/pnas.0709013104. Epub 2007 Nov 26. Proc Natl Acad Sci U S A. 2007. PMID: 18040051 Free PMC article.
-
Reannotation of protein-coding genes based on an improved graphical representation of DNA sequence.J Comput Chem. 2010 Aug;31(11):2126-35. doi: 10.1002/jcc.21500. J Comput Chem. 2010. PMID: 20175214
-
Comparison of RefSeq protein-coding regions in human and vertebrate genomes.BMC Genomics. 2013 Sep 25;14:654. doi: 10.1186/1471-2164-14-654. BMC Genomics. 2013. PMID: 24063302 Free PMC article.
-
Reconsidering proteomic diversity with functional investigation of small ORFs and alternative ORFs.Exp Cell Res. 2020 Aug 1;393(1):112057. doi: 10.1016/j.yexcr.2020.112057. Epub 2020 May 6. Exp Cell Res. 2020. PMID: 32387289 Review.
-
["Matreshka" Genes with Alternative Reading Frames].Genetika. 2016 Feb;52(2):146-63. Genetika. 2016. PMID: 27215029 Review. Russian.
Cited by
-
New genes contribute to genetic and phenotypic novelties in human evolution.Curr Opin Genet Dev. 2014 Dec;29:90-6. doi: 10.1016/j.gde.2014.08.013. Epub 2014 Sep 16. Curr Opin Genet Dev. 2014. PMID: 25218862 Free PMC article. Review.
-
Three polymorphisms of renin-angiotensin system and preeclampsia risk.J Assist Reprod Genet. 2020 Dec;37(12):3121-3142. doi: 10.1007/s10815-020-01971-8. Epub 2020 Nov 23. J Assist Reprod Genet. 2020. PMID: 33230614 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

