Genome-wide landscapes of human local adaptation in Asia
- PMID: 23349834
- PMCID: PMC3551950
- DOI: 10.1371/journal.pone.0054224
Genome-wide landscapes of human local adaptation in Asia
Abstract
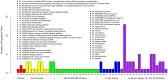
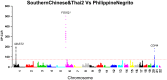
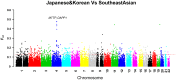
Genetic studies of human local adaptation have been facilitated greatly by recent advances in high-throughput genotyping and sequencing technologies. However, few studies have investigated local adaptation in Asian populations on a genome-wide scale and with a high geographic resolution. In this study, taking advantage of the dense population coverage in Southeast Asia, which is the part of the world least studied in term of natural selection, we depicted genome-wide landscapes of local adaptations in 63 Asian populations representing the majority of linguistic and ethnic groups in Asia. Using genome-wide data analysis, we discovered many genes showing signs of local adaptation or natural selection. Notable examples, such as FOXQ1, MAST2, and CDH4, were found to play a role in hair follicle development and human cancer, signal transduction, and tumor repression, respectively. These showed strong indications of natural selection in Philippine Negritos, a group of aboriginal hunter-gatherers living in the Philippines. MTTP, which has associations with metabolic syndrome, body mass index, and insulin regulation, showed a strong signature of selection in Southeast Asians, including Indonesians. Functional annotation analysis revealed that genes and genetic variants underlying natural selections were generally enriched in the functional category of alternative splicing. Specifically, many genes showing significant difference with respect to allele frequency between northern and southern Asian populations were found to be associated with human height and growth and various immune pathways. In summary, this study contributes to the overall understanding of human local adaptation in Asia and has identified both known and novel signatures of natural selection in the human genome.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

