Cyclic AMP enhances TGFβ responses of breast cancer cells by upregulating TGFβ receptor I expression
- PMID: 23349840
- PMCID: PMC3548810
- DOI: 10.1371/journal.pone.0054261
Cyclic AMP enhances TGFβ responses of breast cancer cells by upregulating TGFβ receptor I expression
Abstract
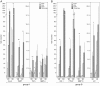
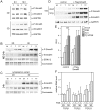
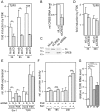
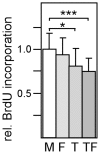
Cellular functions are regulated by complex networks of many different signaling pathways. The TGFβ and cAMP pathways are of particular importance in tumor progression. We analyzed the cross-talk between these pathways in breast cancer cells in 2D and 3D cultures. We found that cAMP potentiated TGFβ-dependent gene expression by enhancing Smad3 phosphorylation. Higher levels of total Smad3, as observed in 3D-cultured cells, blocked this effect. Two Smad3 regulating proteins, YAP (Yes-associated protein) and TβRI (TGFβ receptor 1), were responsive to cAMP. While YAP had little effect on TGFβ-dependent expression and Smad3 phosphorylation, a constitutively active form of TβRI mimicked the cAMP effect on TGFβ signaling. In 3D-cultured cells, which show much higher levels of TβRI and cAMP, TβRI was unresponsive to cAMP. Upregulation of TβRI expression by cAMP was dependent on transcription. A proximal TβRI promoter fragment was moderately, but significantly activated by cAMP suggesting that cAMP increases TβRI expression at least partially by activating TβRI transcription. Neither the cAMP-responsive element binding protein (CREB) nor the TβRI-regulating transcription factor Six1 was required for the cAMP effect. An inhibitor of histone deacetylases alone or together with cAMP increased TβRI expression by a similar extent as cAMP alone suggesting that cAMP may exert its effect by interfering with histone acetylation. Along with an additive stimulatory effect of cAMP and TGFβ on p21 expression an additive inhibitory effect of these agents on proliferation was observed. Finally, we show that mesenchymal stem cells that interact with breast cancer cells can simultaneously activate the cAMP and TGFβ pathways. In summary, these data suggest that combined effects of cAMP and TGFβ, as e.g. induced by mesenchymal stem cells, involve the upregulation of TβRI expression on the transcriptional level, likely due to changes in histone acetylation. As a consequence, cancer cell functions such as proliferation are affected.
Conflict of interest statement
Figures







References
-
- Ikushima H, Miyazono K (2010) TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer 10: 415–424. - PubMed
-
- Schmierer B, Hill CS (2007) TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 8: 970–982. - PubMed
-
- Massague J, Seoane J, Wotton D (2005) Smad transcription factors. Genes Dev 19: 2783–2810. - PubMed
-
- Bierie B, Moses HL (2006) Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer 6: 506–520. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

