NA proteins of influenza A viruses H1N1/2009, H5N1, and H9N2 show differential effects on infection initiation, virus release, and cell-cell fusion
- PMID: 23349854
- PMCID: PMC3551949
- DOI: 10.1371/journal.pone.0054334
NA proteins of influenza A viruses H1N1/2009, H5N1, and H9N2 show differential effects on infection initiation, virus release, and cell-cell fusion
Abstract
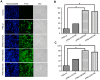
Two surface glycoproteins of influenza virus, haemagglutinin (HA) and neuraminidase (NA), play opposite roles in terms of their interaction with host sialic acid receptors. HA attaches to sialic acid on host cell surface receptors to initiate virus infection while NA removes these sialic acids to facilitate release of progeny virions. This functional opposition requires a balance. To explore what might happen when NA of an influenza virus was replaced by one from another isolate or subtype, in this study, we generated three recombinant influenza A viruses in the background of A/PR/8/34 (PR8) (H1N1) and with NA genes obtained respectively from the 2009 pandemic H1N1 virus, a highly pathogenic avian H5N1 virus, and a lowly pathogenic avian H9N2 virus. These recombinant viruses, rPR8-H1N1NA, rPR8-H5N1NA, and rPR8-H9N2NA, were shown to have similar growth kinetics in cells and pathogenicity in mice. However, much more rPR8-H5N1NA and PR8-wt virions were released from chicken erythrocytes than virions of rPR8-H1N1NA and rPR8-H9N2NA after 1 h. In addition, in MDCK cells, rPR8-H5N1NA and rPR8-H9N2NA infected a higher percentage of cells, and induced cell-cell fusion faster and more extensively than PR8-wt and rPR8-H1N1NA did in the early phase of infection. In conclusion, NA replacement in this study did not affect virus replication kinetics but had different effects on infection initiation, virus release and fusion of infected cells. These phenomena might be partially due to NA proteins' different specificity to α2-3/2-6-sialylated carbohydrate chains, but the exact mechanism remains to be explored.
Conflict of interest statement
Figures
Similar articles
-
Fusion-related host proteins are actively regulated by NA during influenza infection as revealed by quantitative proteomics analysis.PLoS One. 2014 Aug 25;9(8):e105947. doi: 10.1371/journal.pone.0105947. eCollection 2014. PLoS One. 2014. PMID: 25153908 Free PMC article.
-
Effects of different polymerases of avian influenza viruses on the growth and pathogenicity of A/Puerto Rico/8/1934 (H1N1)-derived reassorted viruses.Vet Microbiol. 2014 Jan 10;168(1):41-9. doi: 10.1016/j.vetmic.2013.10.011. Epub 2013 Oct 26. Vet Microbiol. 2014. PMID: 24296300
-
H9N2 Influenza Virus Infections in Human Cells Require a Balance between Neuraminidase Sialidase Activity and Hemagglutinin Receptor Affinity.J Virol. 2020 Aug 31;94(18):e01210-20. doi: 10.1128/JVI.01210-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641475 Free PMC article.
-
The contribution of animal models to the understanding of the host range and virulence of influenza A viruses.Microbes Infect. 2011 May;13(5):502-15. doi: 10.1016/j.micinf.2011.01.014. Epub 2011 Jan 27. Microbes Infect. 2011. PMID: 21276869 Free PMC article. Review.
-
Immune response in influenza virus infection and modulation of immune injury by viral neuraminidase.Virol J. 2023 Aug 28;20(1):193. doi: 10.1186/s12985-023-02164-2. Virol J. 2023. PMID: 37641134 Free PMC article. Review.
Cited by
-
Long-term immunogenicity of an inactivated split-virion 2009 pandemic influenza A H1N1 virus vaccine with or without aluminum adjuvant in mice.Clin Vaccine Immunol. 2015 Mar;22(3):327-35. doi: 10.1128/CVI.00662-14. Epub 2015 Jan 14. Clin Vaccine Immunol. 2015. PMID: 25589552 Free PMC article.
-
The Contribution of Viral Proteins to the Synergy of Influenza and Bacterial Co-Infection.Viruses. 2022 May 16;14(5):1064. doi: 10.3390/v14051064. Viruses. 2022. PMID: 35632805 Free PMC article. Review.
-
Competitive Cooperation of Hemagglutinin and Neuraminidase during Influenza A Virus Entry.Viruses. 2019 May 20;11(5):458. doi: 10.3390/v11050458. Viruses. 2019. PMID: 31137516 Free PMC article. Review.
-
Fusion-related host proteins are actively regulated by NA during influenza infection as revealed by quantitative proteomics analysis.PLoS One. 2014 Aug 25;9(8):e105947. doi: 10.1371/journal.pone.0105947. eCollection 2014. PLoS One. 2014. PMID: 25153908 Free PMC article.
-
A new role of neuraminidase (NA) in the influenza virus life cycle: implication for developing NA inhibitors with novel mechanism of action.Rev Med Virol. 2016 Jul;26(4):242-50. doi: 10.1002/rmv.1879. Epub 2016 Apr 8. Rev Med Virol. 2016. PMID: 27061123 Free PMC article. Review.
References
-
- Horimoto T, Kawaoka Y (2005) Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3: 591–600. - PubMed
-
- Morens DM, Fauci AS (2007) The 1918 influenza pandemic: insights for the 21st century. J Infect Dis 195: 1018–1028. - PubMed
-
- WHO website. Available: http://www.who.int/influenza/human_animal_interface/EN_GIP_20120810Cumul.... Accessed 2012 Aug 10.
-
- Girard MP, Tam JS, Assossou OM, Kieny MP (2010) The 2009 A (H1N1) influenza virus pandemic: A review. Vaccine 28: 4895–4902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

