In vitro secondary structure of the genomic RNA of satellite tobacco mosaic virus
- PMID: 23349871
- PMCID: PMC3551766
- DOI: 10.1371/journal.pone.0054384
In vitro secondary structure of the genomic RNA of satellite tobacco mosaic virus
Abstract
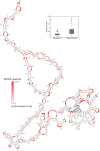
Satellite tobacco mosaic virus (STMV) is a T = 1 icosahedral virus with a single-stranded RNA genome. It is widely accepted that the RNA genome plays an important structural role during assembly of the STMV virion. While the encapsidated form of the RNA has been extensively studied, less is known about the structure of the free RNA, aside from a purported tRNA-like structure at the 3' end. Here we use selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) analysis to examine the secondary structure of in vitro transcribed STMV RNA. The predicted secondary structure is unusual in the sense that it is highly extended, which could be significant for protecting the RNA from degradation. The SHAPE data are also consistent with the previously predicted tRNA-like fold at the 3' end of the molecule, which is also known to hinder degradation. Our data are not consistent with the secondary structure proposed for the encapsidated RNA by Schroeder et al., suggesting that, if the Schroeder structure is correct, either the RNA is packaged as it emerges from the replication complex, or the RNA undergoes extensive refolding upon encapsidation. We also consider the alternative, i.e., that the structure of the encapsidated STMV RNA might be the same as the in vitro structure presented here, and we examine how this structure might be organized in the virus. This possibility is not rigorously ruled out by the available data, so it remains open to examination by experiment.
Conflict of interest statement
Figures









References
-
- Dodds JA (1998) Satellite tobacco mosaic virus. Annual Review of Phytopathology 36: 295–310. - PubMed
-
- Schneemann A (2006) The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol 60: 51–67. - PubMed
-
- Gossele V, Fache I, Meulewaeter F, Cornelissen M, Metzlaff M (2002) SVISS - a novel transient gene silencing system for gene function discovery and validation in tobacco plants. Plant Journal 32: 859–866. - PubMed
-
- Larson SB, Day J, Greenwood A, McPherson A (1998) Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J Mol Biol 277: 37–59. - PubMed
-
- Larson SB, McPherson A (2001) Satellite tobacco mosaic virus RNA: Structure and implications for assembly. Curr Opinion Struct Biol 11: 59–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

