Detection of copy number variants reveals association of cilia genes with neural tube defects
- PMID: 23349908
- PMCID: PMC3547935
- DOI: 10.1371/journal.pone.0054492
Detection of copy number variants reveals association of cilia genes with neural tube defects
Abstract
Background: Neural tube defects (NTDs) are one of the most common birth defects caused by a combination of genetic and environmental factors. Currently, little is known about the genetic basis of NTDs although up to 70% of human NTDs were reported to be attributed to genetic factors. Here we performed genome-wide copy number variants (CNVs) detection in a cohort of Chinese NTD patients in order to exam the potential role of CNVs in the pathogenesis of NTDs.
Methods: The genomic DNA from eighty-five NTD cases and seventy-five matched normal controls were subjected for whole genome CNVs analysis. Non-DGV (the Database of Genomic Variants) CNVs from each group were further analyzed for their associations with NTDs. Gene content in non-DGV CNVs as well as participating pathways were examined.
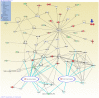
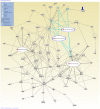
Results: Fifty-five and twenty-six non-DGV CNVs were detected in cases and controls respectively. Among them, forty and nineteen CNVs involve genes (genic CNV). Significantly more non-DGV CNVs and non-DGV genic CNVs were detected in NTD patients than in control (41.2% vs. 25.3%, p<0.05 and 37.6% vs. 20%, p<0.05). Non-DGV genic CNVs are associated with a 2.65-fold increased risk for NTDs (95% CI: 1.24-5.87). Interestingly, there are 41 cilia genes involved in non-DGV CNVs from NTD patients which is significantly enriched in cases compared with that in controls (24.7% vs. 9.3%, p<0.05), corresponding with a 3.19-fold increased risk for NTDs (95% CI: 1.27-8.01). Pathway analyses further suggested that two ciliogenesis pathways, tight junction and protein kinase A signaling, are top canonical pathways implicated in NTD-specific CNVs, and these two novel pathways interact with known NTD pathways.
Conclusions: Evidence from the genome-wide CNV study suggests that genic CNVs, particularly ciliogenic CNVs are associated with NTDs and two ciliogenesis pathways, tight junction and protein kinase A signaling, are potential pathways involved in NTD pathogenesis.
Conflict of interest statement
Figures


References
-
- Botto LD, Moore CA, Khoury MJ, Erickson JD (1999) Neural-tube defects. N Engl J Med 341: 1509–1519. - PubMed
-
- Feuchtbaum LB, Currier RJ, Riggle S, Roberson M, Lorey FW, et al. (1999) Neural tube defect prevalence in California (1990–1994): eliciting patterns by type of defect and maternal race/ethnicity. Genet Test 3: 265–272. - PubMed
-
- KZ X (1989) The epidemiology of neural tube defects in China. Chin J Med 69: 189–191. - PubMed
-
- Li Z, Ren A, Zhang L, Ye R, Li S, et al. (2006) Extremely high prevalence of neural tube defects in a 4-county area in Shanxi Province, China. Birth Defects Res A Clin Mol Teratol 76: 237–240. - PubMed
-
- Gu X, Lin L, Zheng X, Zhang T, Song X, et al. (2007) High prevalence of NTDs in Shanxi Province: a combined epidemiological approach. Birth Defects Res A Clin Mol Teratol 79: 702–707. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

