A multiplex PCR for the simultaneous detection and genotyping of the Echinococcus granulosus complex
- PMID: 23350011
- PMCID: PMC3547860
- DOI: 10.1371/journal.pntd.0002017
A multiplex PCR for the simultaneous detection and genotyping of the Echinococcus granulosus complex
Abstract
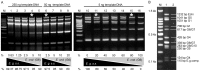
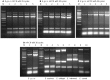
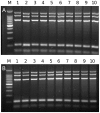
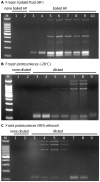
Echinococcus granulosus is characterized by high intra-specific variability (genotypes G1-G10) and according to the new molecular phylogeny of the genus Echinococcus, the E. granulosus complex has been divided into E. granulosus sensu stricto (G1-G3), E. equinus (G4), E. ortleppi (G5), and E. canadensis (G6-G10). The molecular characterization of E. granulosus isolates is fundamental to understand the spatio-temporal epidemiology of this complex in many endemic areas with the simultaneous occurrence of different Echinococcus species and genotypes. To simplify the genotyping of the E. granulosus complex we developed a single-tube multiplex PCR (mPCR) allowing three levels of discrimination: (i) Echinococcus genus, (ii) E. granulosus complex in common, and (iii) the specific genotype within the E. granulosus complex. The methodology was established with known DNA samples of the different strains/genotypes, confirmed on 42 already genotyped samples (Spain: 22 and Bulgaria: 20) and then successfully applied on 153 unknown samples (Tunisia: 114, Algeria: 26 and Argentina: 13). The sensitivity threshold of the mPCR was found to be 5 ng Echinoccoccus DNA in a mixture of up to 1 µg of foreign DNA and the specificity was 100% when template DNA from closely related members of the genus Taenia was used. Additionally to DNA samples, the mPCR can be carried out directly on boiled hydatid fluid or on alkaline-lysed frozen or fixed protoscoleces, thus avoiding classical DNA extractions. However, when using Echinococcus eggs obtained from fecal samples of infected dogs, the sensitivity of the mPCR was low (<40%). Thus, except for copro analysis, the mPCR described here has a high potential for a worldwide application in large-scale molecular epidemiological studies on the Echinococcus genus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A novel PCR-RFLP assay for molecular characterization of Echinococcus granulosus sensu lato and closely related species in developing countries.Parasitol Res. 2016 Oct;115(10):3817-24. doi: 10.1007/s00436-016-5143-x. Epub 2016 May 25. Parasitol Res. 2016. PMID: 27225001
-
The optimum cut-off value to differentiate Echinococcus granulosus sensu stricto from other species of E. granulosus sensu lato using larval rostellar hook morphometry.J Helminthol. 2015 Jan;89(1):1-8. doi: 10.1017/S0022149X13000473. Epub 2013 Jul 10. J Helminthol. 2015. PMID: 23842071
-
Echinococcus granulosus sensu lato genotypes infecting humans--review of current knowledge.Int J Parasitol. 2014 Jan;44(1):9-18. doi: 10.1016/j.ijpara.2013.08.008. Epub 2013 Nov 19. Int J Parasitol. 2014. PMID: 24269720 Review.
-
Molecular characterization of Echinococcus granulosus sensu stricto and Echinococcus canadensis in humans and livestock from Algeria.Parasitol Res. 2016 Jun;115(6):2423-31. doi: 10.1007/s00436-016-4994-5. Epub 2016 Mar 28. Parasitol Res. 2016. PMID: 27021186
-
Species and genotypes belonging to Echinococcus granulosus sensu lato complex causing human cystic echinococcosis in Europe (2000-2021): a systematic review.Parasit Vectors. 2022 Mar 28;15(1):109. doi: 10.1186/s13071-022-05197-8. Parasit Vectors. 2022. PMID: 35346335 Free PMC article.
Cited by
-
Human Cystic Echinococcosis in the Nalut District of Western Libya: A Clinico-epidemiological Study.Trop Med Health. 2014 Dec;42(4):177-84. doi: 10.2149/tmh.2014-16. Epub 2014 Sep 17. Trop Med Health. 2014. PMID: 25589882 Free PMC article.
-
Genetic survey of cystic echinococcosis in farm animals in Oman.Trop Anim Health Prod. 2020 Jan;52(1):331-337. doi: 10.1007/s11250-019-02019-5. Epub 2019 Jul 23. Trop Anim Health Prod. 2020. PMID: 31338730
-
Update on the genetic diversity and population structure of Echinococcus granulosus in Gansu Province, Tibet Autonomous Region, and Xinjiang Uygur Autonomous Region, Western China, inferred from mitochondrial cox1, nad1, and nad5 sequences.Parasitol Res. 2023 May;122(5):1107-1126. doi: 10.1007/s00436-023-07811-9. Epub 2023 Mar 18. Parasitol Res. 2023. PMID: 36933066
-
Occurrence and genetic characterization of Echinococcus granulosus sensu lato from domestic animals in Central Iran.BMC Vet Res. 2022 Jan 7;18(1):22. doi: 10.1186/s12917-021-03131-1. BMC Vet Res. 2022. PMID: 34996460 Free PMC article.
-
Identification of Echinococcus granulosus strains using polymerase chain reaction-restriction fragment length polymorphism amongst livestock in Moroto district, Uganda.Onderstepoort J Vet Res. 2016 Jul 29;83(1):e1-7. doi: 10.4102/ojvr.v83i1.1068. Onderstepoort J Vet Res. 2016. PMID: 27543147 Free PMC article.
References
-
- Hansen B (1991) New York City epidemics and history for the public. In: Harden VA, Risse GB, editors. AIDS and the historian. Bethesda: National Institutes of Health. pp. 21–28.
-
- Xiao N, Qiu JM, Nakao M, Li TY, Yang W, et al. (2005) Echinococcus shiquicus n. sp., a taeniid cestod from Tibetan fox and plateau pika in China. Int J Parasitol 35: 693–701. - PubMed
-
- Hüttner M, Nakao M, Wassermann T, Siefert L, Boomker JD, et al. (2008) Genetic characterization and phylogenetic position of Echinococcus felidis (Cestoda: Taeniidae) from the African lion. Int J Parasitol 7: 861–868. - PubMed
-
- McManus DP, Thompson RC (2003) Molecular epidemiology of cystic echinococcosis. Parasitology 127: 37–51. - PubMed
-
- Thompson RC, McManus DP (2002) Towards a taxonomic revision of the genus Echinococcus . Trends Parasitol 18: 452–457. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

