Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants
- PMID: 23350908
- PMCID: PMC3576282
- DOI: 10.1186/1471-2164-14-56
Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants
Abstract
Background: Evidence has been assembled to suggest synonymous codon usage bias (SCUB) has close relationship with intron. However, the relationship (if any) between SCUB and intron number as well as exon position is at present rather unclear.
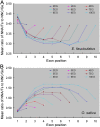
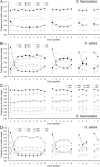
Results: To explore this relationship, the sequences of a set of genes containing between zero and nine introns was extracted from the published genome sequences of three algal species, one moss, one fern and six angiosperms (three monocotyledonous species and three dicotyledonous species). In the algal genomes, the frequency of synonymous codons of the form NNG/NNC (codons with G and C at the third position) was positively related to intron number, but that of NNA/NNT was inversely correlated; the opposite was the case in the land plant genomes. The frequency of NNC/NNG was higher and that of NNA/NNT lower in two terminal exons than in the interstitial exons in the land plant genes, but the rule showed to be opposite in the algal genes. SCUB patterns in the interstitial and two terminal exons mirror the different evolutionary relationships between these plant species, while the first exon shows the highest level of conservation is therefore concluded to be the one which experiences the heaviest selection pressure. The phenomenon of SCUB may also be related to DNA methylation induced conversion of CG to AT.
Conclusions: These data provide some evidence of linkage between SCUB, the evolution of introns and DNA methylation, which brings about a new perspective for understanding how genomic variation is created during plant evolution.
Figures





Similar articles
-
The Shift in Synonymous Codon Usage Reveals Similar Genomic Variation during Domestication of Asian and African Rice.Int J Mol Sci. 2022 Oct 25;23(21):12860. doi: 10.3390/ijms232112860. Int J Mol Sci. 2022. PMID: 36361651 Free PMC article.
-
Synonymous codon usage bias in plant mitochondrial genes is associated with intron number and mirrors species evolution.PLoS One. 2015 Jun 25;10(6):e0131508. doi: 10.1371/journal.pone.0131508. eCollection 2015. PLoS One. 2015. PMID: 26110418 Free PMC article.
-
Synonymous Codon Usage Bias in the Plastid Genome is Unrelated to Gene Structure and Shows Evolutionary Heterogeneity.Evol Bioinform Online. 2015 Apr 7;11:65-77. doi: 10.4137/EBO.S22566. eCollection 2015. Evol Bioinform Online. 2015. PMID: 25922569 Free PMC article.
-
Analysis of evolution of exon-intron structure of eukaryotic genes.Brief Bioinform. 2005 Jun;6(2):118-34. doi: 10.1093/bib/6.2.118. Brief Bioinform. 2005. PMID: 15975222 Review.
-
Evolution of the intron-exon structure of eukaryotic genes.Curr Opin Genet Dev. 1995 Dec;5(6):774-8. doi: 10.1016/0959-437x(95)80010-3. Curr Opin Genet Dev. 1995. PMID: 8745076 Review.
Cited by
-
Alteration of synonymous codon usage bias accompanies polyploidization in wheat.Front Genet. 2022 Oct 14;13:979902. doi: 10.3389/fgene.2022.979902. eCollection 2022. Front Genet. 2022. PMID: 36313462 Free PMC article.
-
Comparative Analyses of Complete Chloroplast Genomes of Microula sikkimensis and Related Species of Boraginaceae.Genes (Basel). 2024 Feb 10;15(2):226. doi: 10.3390/genes15020226. Genes (Basel). 2024. PMID: 38397215 Free PMC article.
-
The Shift in Synonymous Codon Usage Reveals Similar Genomic Variation during Domestication of Asian and African Rice.Int J Mol Sci. 2022 Oct 25;23(21):12860. doi: 10.3390/ijms232112860. Int J Mol Sci. 2022. PMID: 36361651 Free PMC article.
-
Introns Structure Patterns of Variation in Nucleotide Composition in Arabidopsis thaliana and Rice Protein-Coding Genes.Genome Biol Evol. 2015 Oct 7;7(10):2913-28. doi: 10.1093/gbe/evv189. Genome Biol Evol. 2015. PMID: 26450849 Free PMC article.
-
Chloroplast Genome Variation and Phylogenetic Relationships of Autochthonous Varieties of Vitis vinifera from the Don Valley.Int J Mol Sci. 2024 Sep 14;25(18):9928. doi: 10.3390/ijms25189928. Int J Mol Sci. 2024. PMID: 39337416 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

