Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders
- PMID: 23352160
- PMCID: PMC3613849
- DOI: 10.1016/j.neuron.2012.12.029
Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders
Abstract
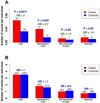
To characterize the role of rare complete human knockouts in autism spectrum disorders (ASDs), we identify genes with homozygous or compound heterozygous loss-of-function (LoF) variants (defined as nonsense and essential splice sites) from exome sequencing of 933 cases and 869 controls. We identify a 2-fold increase in complete knockouts of autosomal genes with low rates of LoF variation (≤ 5% frequency) in cases and estimate a 3% contribution to ASD risk by these events, confirming this observation in an independent set of 563 probands and 4,605 controls. Outside the pseudoautosomal regions on the X chromosome, we similarly observe a significant 1.5-fold increase in rare hemizygous knockouts in males, contributing to another 2% of ASDs in males. Taken together, these results provide compelling evidence that rare autosomal and X chromosome complete gene knockouts are important inherited risk factors for ASD.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Rare inherited variation in autism: beginning to see the forest and a few trees.Neuron. 2013 Jan 23;77(2):209-11. doi: 10.1016/j.neuron.2013.01.010. Neuron. 2013. PMID: 23352155 Free PMC article.
References
-
- Prevalence of autism spectrum disorders--Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. - PubMed
-
- Celestino-Soper PB, Shaw CA, Sanders SJ, Li J, Murtha MT, Ercan-Sencicek AG, Davis L, Thomson S, Gambin T, Chinault AC, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum Mol Genet. 2011;20:4360–4370. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U24 MH081810/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 MH089004/MH/NIMH NIH HHS/United States
- R01 MH057881/MH/NIMH NIH HHS/United States
- HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- R01 MH089482/MH/NIMH NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 MH089208/MH/NIMH NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-103010/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P50HD055751/HD/NICHD NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 MH061009/MH/NIMH NIH HHS/United States
- U24MH068457/MH/NIMH NIH HHS/United States
- 1U24MH081810/MH/NIMH NIH HHS/United States
- R01MH061009/MH/NIMH NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01MH089175/MH/NIMH NIH HHS/United States
- P50 HD055751/HD/NICHD NIH HHS/United States
- HL-102926/HL/NHLBI NIH HHS/United States
- R01 MH089025/MH/NIMH NIH HHS/United States
- R01MH089004/MH/NIMH NIH HHS/United States
- R01MH089208/MH/NIMH NIH HHS/United States
- R01 MH089175/MH/NIMH NIH HHS/United States
- R01MH057881/MH/NIMH NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- HL-102923/HL/NHLBI NIH HHS/United States
- R01MH089025/MH/NIMH NIH HHS/United States
- R01MH089482/MH/NIMH NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

