From systems to structure: bridging networks and mechanism
- PMID: 23352243
- PMCID: PMC3558917
- DOI: 10.1016/j.molcel.2013.01.003
From systems to structure: bridging networks and mechanism
Abstract
There is a wide gap between the generation of large-scale biological data sets and more-detailed, structural and mechanistic studies. However, recent studies that explicitly combine data from systems and structural biological approaches are having a profound effect on our ability to predict how mutations and small molecules affect atomic-level mechanisms, disrupt systems-level networks, and ultimately lead to changes in organismal fitness. In fact, we argue that a shared framework for analysis of nonadditive genetic and thermodynamic responses to perturbations will accelerate the integration of reductionist and global approaches. A stronger bridge between these two areas will allow for a deeper and more-complete understanding of complex biological phenomenon and ultimately provide needed breakthroughs in biomedical research.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
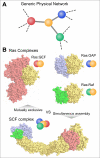



References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
-
- Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. - PubMed
Publication types
MeSH terms
Grants and funding
- GM082250/GM/NIGMS NIH HHS/United States
- GM081879/GM/NIGMS NIH HHS/United States
- GM084279/GM/NIGMS NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- GM084448/GM/NIGMS NIH HHS/United States
- R01 GM084279/GM/NIGMS NIH HHS/United States
- P01 AI091575/AI/NIAID NIH HHS/United States
- AI090935/AI/NIAID NIH HHS/United States
- R01 GM098101/GM/NIGMS NIH HHS/United States
- GM098101/GM/NIGMS NIH HHS/United States
- DP5OD009180/OD/NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01 GM084448/GM/NIGMS NIH HHS/United States
- DP5 OD009180/OD/NIH HHS/United States
- AI091575/AI/NIAID NIH HHS/United States
- GM078360/GM/NIGMS NIH HHS/United States
- R01 GM078360/GM/NIGMS NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

