Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability
- PMID: 23352259
- PMCID: PMC3567272
- DOI: 10.1016/j.ajhg.2012.12.014
Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability
Abstract
Syndromes associated with multiple mtDNA deletions are due to different molecular defects that can result in a wide spectrum of predominantly adult-onset clinical presentations, ranging from progressive external ophthalmoplegia (PEO) to multisystemic disorders of variable severity. The autosomal-dominant form of PEO is genetically heterogeneous. Recently, causative mutations have been reported in several nuclear genes that encode proteins of the mtDNA replisome machinery (POLG, POLG2, and C10orf2) or that are involved in pathways for the synthesis of deoxyribonuclotides (ANT1 and RRM2B). Despite these findings, putative mutations remain unknown in half of the subjects with PEO. We report the identification, by exome sequencing, of mutations in DNA2 in adult-onset individuals with a form of mitochondrial myopathy featuring instability of muscle mtDNA. DNA2 encodes a helicase/nuclease family member that is most likely involved in mtDNA replication, as well as in the long-patch base-excision repair (LP-BER) pathway. In vitro biochemical analysis of purified mutant proteins revealed a severe impairment of nuclease, helicase, and ATPase activities. These results implicate human DNA2 and the LP-BER pathway in the pathogenesis of adult-onset disorders of mtDNA maintenance.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
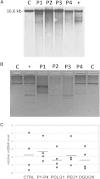
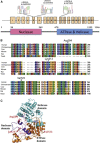

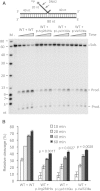
References
-
- Wanrooij S., Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta. 2010;1797:1378–1388. - PubMed
-
- Virgilio R., Ronchi D., Hadjigeorgiou G.M., Bordoni A., Saladino F., Moggio M., Adobbati L., Kafetsouli D., Tsironi E., Previtali S. Novel Twinkle (PEO1) gene mutations in mendelian progressive external ophthalmoplegia. J. Neurol. 2008;255:1384–1391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

