Global protein phosphorylation dynamics during deoxynivalenol-induced ribotoxic stress response in the macrophage
- PMID: 23352502
- PMCID: PMC4041276
- DOI: 10.1016/j.taap.2013.01.007
Global protein phosphorylation dynamics during deoxynivalenol-induced ribotoxic stress response in the macrophage
Abstract
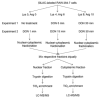
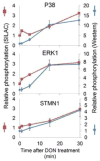
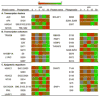
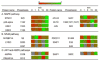
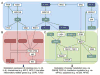
Deoxynivalenol (DON), a trichothecene mycotoxin produced by Fusarium that commonly contaminates food, is capable of activating mononuclear phagocytes of the innate immune system via a process termed the ribotoxic stress response (RSR). To encapture global signaling events mediating RSR, we quantified the early temporal (≤30min) phosphoproteome changes that occurred in RAW 264.7 murine macrophage during exposure to a toxicologically relevant concentration of DON (250ng/mL). Large-scale phosphoproteomic analysis employing stable isotope labeling of amino acids in cell culture (SILAC) in conjunction with titanium dioxide chromatography revealed that DON significantly upregulated or downregulated phosphorylation of 188 proteins at both known and yet-to-be functionally characterized phosphosites. DON-induced RSR is extremely complex and goes far beyond its prior known capacity to inhibit translation and activate MAPKs. Transcriptional regulation was the main target during early DON-induced RSR, covering over 20% of the altered phosphoproteins as indicated by Gene Ontology annotation and including transcription factors/cofactors and epigenetic modulators. Other biological processes impacted included cell cycle, RNA processing, translation, ribosome biogenesis, monocyte differentiation and cytoskeleton organization. Some of these processes could be mediated by signaling networks involving MAPK-, NFκB-, AKT- and AMPK-linked pathways. Fuzzy c-means clustering revealed that DON-regulated phosphosites could be discretely classified with regard to the kinetics of phosphorylation/dephosphorylation. The cellular response networks identified provide a template for further exploration of the mechanisms of trichothecenemycotoxins and other ribotoxins, and ultimately, could contribute to improved mechanism-based human health risk assessment.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

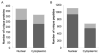



Similar articles
-
Early phosphoproteomic changes in the mouse spleen during deoxynivalenol-induced ribotoxic stress.Toxicol Sci. 2013 Sep;135(1):129-43. doi: 10.1093/toxsci/kft145. Epub 2013 Jun 29. Toxicol Sci. 2013. PMID: 23811945 Free PMC article.
-
Dynamic changes in ribosome-associated proteome and phosphoproteome during deoxynivalenol-induced translation inhibition and ribotoxic stress.Toxicol Sci. 2014 Mar;138(1):217-33. doi: 10.1093/toxsci/kft270. Epub 2013 Nov 27. Toxicol Sci. 2014. PMID: 24284785 Free PMC article.
-
Ribotoxic stress response to the trichothecene deoxynivalenol in the macrophage involves the SRC family kinase Hck.Toxicol Sci. 2005 Jun;85(2):916-26. doi: 10.1093/toxsci/kfi146. Epub 2005 Mar 16. Toxicol Sci. 2005. PMID: 15772366
-
Deoxynivalenol-induced proinflammatory gene expression: mechanisms and pathological sequelae.Toxins (Basel). 2010 Jun;2(6):1300-17. doi: 10.3390/toxins2061300. Epub 2010 Jun 1. Toxins (Basel). 2010. PMID: 22069639 Free PMC article. Review.
-
Mechanisms of deoxynivalenol-induced gene expression and apoptosis.Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008 Sep;25(9):1128-40. doi: 10.1080/02652030802056626. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008. PMID: 19238623 Free PMC article. Review.
Cited by
-
TaFROG Encodes a Pooideae Orphan Protein That Interacts with SnRK1 and Enhances Resistance to the Mycotoxigenic Fungus Fusarium graminearum.Plant Physiol. 2015 Dec;169(4):2895-906. doi: 10.1104/pp.15.01056. Epub 2015 Oct 27. Plant Physiol. 2015. PMID: 26508775 Free PMC article.
-
Mercury alters B-cell protein phosphorylation profiles.J Proteome Res. 2014 Feb 7;13(2):496-505. doi: 10.1021/pr400657k. Epub 2013 Dec 4. J Proteome Res. 2014. PMID: 24224561 Free PMC article.
-
Exploring the dermotoxicity of the mycotoxin deoxynivalenol: combined morphologic and proteomic profiling of human epidermal cells reveals alteration of lipid biosynthesis machinery and membrane structural integrity relevant for skin barrier function.Arch Toxicol. 2021 Jun;95(6):2201-2221. doi: 10.1007/s00204-021-03042-y. Epub 2021 Apr 23. Arch Toxicol. 2021. PMID: 33890134 Free PMC article.
-
Phosphoproteome Analysis Reveals the Molecular Mechanisms Underlying Deoxynivalenol-Induced Intestinal Toxicity in IPEC-J2 Cells.Toxins (Basel). 2016 Sep 22;8(10):270. doi: 10.3390/toxins8100270. Toxins (Basel). 2016. PMID: 27669298 Free PMC article.
-
Endoplasmic Reticulum Adaptation and Autophagic Competence Shape Response to Fluid Shear Stress in T24 Bladder Cancer Cells.Front Pharmacol. 2021 May 3;12:647350. doi: 10.3389/fphar.2021.647350. eCollection 2021. Front Pharmacol. 2021. PMID: 34012396 Free PMC article.
References
-
- Andersen ME, Krewski D. Toxicity testing in the 21st century: bringing the vision to life. Toxicol Sci. 2009;107:324–330. - PubMed
-
- Azcona-Olivera JI, Ouyang Y, Murtha J, Chu FS, Pestka JJ. Induction of cytokine mRNAs in mice after oral exposure to the trichothecene vomitoxin (deoxynivalenol): relationship to toxin distribution and protein synthesis inhibition. Toxicology and applied pharmacology. 1995;133:109–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

