A new structural paradigm in copper resistance in Streptococcus pneumoniae
- PMID: 23354287
- PMCID: PMC3578076
- DOI: 10.1038/nchembio.1168
A new structural paradigm in copper resistance in Streptococcus pneumoniae
Abstract
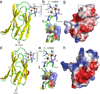
Copper resistance has emerged as an important virulence determinant of microbial pathogens. In Streptococcus pneumoniae, copper resistance is mediated by the copper-responsive repressor CopY, CupA and the copper-effluxing P(1B)-type ATPase CopA. We show here that CupA is a previously uncharacterized cell membrane-anchored Cu(I) chaperone and that a Cu(I) binding-competent, membrane-localized CupA is obligatory for copper resistance. The crystal structures of the soluble domain of CupA and the N-terminal metal-binding domain (MBD) of CopA (CopA(MBD)) reveal isostructural cupredoxin-like folds that each harbor a binuclear Cu(I) cluster unprecedented in bacterial copper trafficking. NMR studies reveal unidirectional Cu(I) transfer from the low-affinity site on the soluble domain of CupA to the high-affinity site of CopA(MBD). However, copper binding by CopA(MBD) is not essential for cellular copper resistance, consistent with a primary role of CupA in cytoplasmic Cu(I) sequestration and/or direct delivery to the transmembrane site of CopA for cellular efflux.
Figures




References
-
- Pena MM, Lee J, Thiele DJ. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 1999;129:1251–1260. - PubMed
-
- Halliwell B, Gutteridge JM. The importance of free radicals and catalytic metal ions in human diseases. Mol. Aspects Med. 1985;8:89–193. - PubMed
-
- Percival SS. Copper and immunity. Am. J. Clin. Nutr. 1998;67:1064S–1068S. - PubMed
-
- Braun V, Hantke K. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 2011;15:328–334. - PubMed
-
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

