Cytochrome P450 initiates degradation of cis-dichloroethene by Polaromonas sp. strain JS666
- PMID: 23354711
- PMCID: PMC3623248
- DOI: 10.1128/AEM.03445-12
Cytochrome P450 initiates degradation of cis-dichloroethene by Polaromonas sp. strain JS666
Abstract
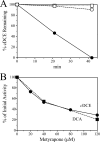
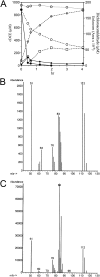
Polaromonas sp. strain JS666 grows on cis-1,2-dichoroethene (cDCE) as the sole carbon and energy source under aerobic conditions, but the degradation mechanism and the enzymes involved are unknown. In this study, we established the complete pathway for cDCE degradation through heterologous gene expression, inhibition studies, enzyme assays, and analysis of intermediates. Several lines of evidence indicate that a cytochrome P450 monooxygenase catalyzes the initial step of cDCE degradation. Both the transient accumulation of dichloroacetaldehyde in cDCE-degrading cultures and dichloroacetaldehyde dehydrogenase activities in cell extracts of JS666 support a pathway for degradation of cDCE through dichloroacetaldehyde. The mechanism minimizes the formation of cDCE epoxide. The molecular phylogeny of the cytochrome P450 gene and the organization of neighboring genes suggest that the cDCE degradation pathway recently evolved in a progenitor capable of degrading 1,2-dichloroethane either by the recruitment of the cytochrome P450 monooxygenase gene from an alkane catabolic pathway or by selection for variants of the P450 in a preexisting 1,2-dichloroethane catabolic pathway. The results presented here add yet another role to the broad array of productive reactions catalyzed by cytochrome P450 enzymes.
Figures






Similar articles
-
Proteomic and transcriptomic analyses reveal genes upregulated by cis-dichloroethene in Polaromonas sp. strain JS666.Appl Environ Microbiol. 2009 Jun;75(11):3733-44. doi: 10.1128/AEM.00031-09. Epub 2009 Apr 10. Appl Environ Microbiol. 2009. PMID: 19363075 Free PMC article.
-
Branched pathways in the degradation of cDCE by cytochrome P450 in Polaromonas sp. JS666.Sci Total Environ. 2017 Dec 15;605-606:99-105. doi: 10.1016/j.scitotenv.2017.06.166. Epub 2017 Jun 26. Sci Total Environ. 2017. PMID: 28662431
-
Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium.Appl Environ Microbiol. 2002 Jun;68(6):2726-30. doi: 10.1128/AEM.68.6.2726-2730.2002. Appl Environ Microbiol. 2002. PMID: 12039726 Free PMC article.
-
Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution.FEMS Microbiol Rev. 2010 Jul;34(4):445-75. doi: 10.1111/j.1574-6976.2010.00210.x. Epub 2010 Jan 8. FEMS Microbiol Rev. 2010. PMID: 20146755 Review.
-
Diversification of P450 genes during land plant evolution.Annu Rev Plant Biol. 2010;61:291-315. doi: 10.1146/annurev-arplant-042809-112305. Annu Rev Plant Biol. 2010. PMID: 20192745 Review.
Cited by
-
A New Catabolic Plasmid in Xanthobacter and Starkeya spp. from a 1,2-Dichloroethane-Contaminated Site.Appl Environ Microbiol. 2016 Aug 15;82(17):5298-308. doi: 10.1128/AEM.01373-16. Print 2016 Sep 1. Appl Environ Microbiol. 2016. PMID: 27342553 Free PMC article.
-
1,2-DCA biodegradation potential of an aquifer assessed in situ and in aerobic and anaerobic microcosms.Environ Microbiome. 2024 Dec 18;19(1):106. doi: 10.1186/s40793-024-00650-w. Environ Microbiome. 2024. PMID: 39696724 Free PMC article.
-
Enzyme activity and gene expression profiles of Xanthobacter autotrophicus GJ10 during aerobic biodegradation of 1,2-dichloroethane.World J Microbiol Biotechnol. 2015 Aug;31(8):1211-6. doi: 10.1007/s11274-015-1868-4. Epub 2015 May 10. World J Microbiol Biotechnol. 2015. PMID: 25957483
-
Microbial degradation of chloroethenes: a review.Environ Sci Pollut Res Int. 2017 May;24(15):13262-13283. doi: 10.1007/s11356-017-8867-y. Epub 2017 Apr 5. Environ Sci Pollut Res Int. 2017. PMID: 28378313 Review.
-
Genomic analysis of Acinetobacter pittii CEP14 reveals its extensive biodegradation capabilities, including cometabolic degradation of cis-1,2-dichloroethene.Antonie Van Leeuwenhoek. 2022 Aug;115(8):1041-1057. doi: 10.1007/s10482-022-01752-6. Epub 2022 Jun 15. Antonie Van Leeuwenhoek. 2022. PMID: 35701646
References
-
- Agency for Toxic Substances and Disease Registry 2011. ATSDR 2011 substance priority list. Agency for Toxic Substances and Disease Registry, U.S. Department of Health and Human Services, Atlanta, GA
-
- Maymó-Gatell X, Chien Y, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

