Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes
- PMID: 23355396
- PMCID: PMC3576511
- DOI: 10.1101/gad.206458.112
Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes
Abstract
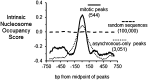
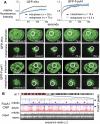
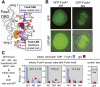
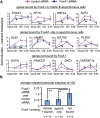
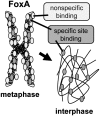
While most transcription factors exit the chromatin during mitosis and the genome becomes silent, a subset of factors remains and "bookmarks" genes for rapid reactivation as cells progress through the cell cycle. However, it is unknown whether such bookmarking factors bind to chromatin similarly in mitosis and how different binding capacities among them relate to function. We compared a diverse set of transcription factors involved in liver differentiation and found markedly different extents of mitotic chromosome binding. Among them, the pioneer factor FoxA1 exhibits the greatest extent of mitotic chromosome binding. Genomically, ~15% of the FoxA1 interphase target sites are bound in mitosis, including at genes that are important for liver differentiation. Biophysical, genome mapping, and mutagenesis studies of FoxA1 reveals two different modes of binding to mitotic chromatin. Specific binding in mitosis occurs at sites that continue to be bound from interphase. Nonspecific binding in mitosis occurs across the chromosome due to the intrinsic chromatin affinity of FoxA1. Both specific and nonspecific binding contribute to timely reactivation of target genes post-mitosis. These studies reveal an unexpected diversity in the mechanisms by which transcription factors help retain cell identity during mitosis.
Figures


Comment in
-
Pioneering barren land: mitotic bookmarking by transcription factors.Dev Cell. 2013 Feb 25;24(4):342-4. doi: 10.1016/j.devcel.2013.02.005. Dev Cell. 2013. PMID: 23449470
References
-
- Agard DA 1984. Optical sectioning microscopy: Cellular architecture in three dimensions. Annu Rev Biophys Bioeng 13: 191–219 - PubMed
-
- Bossard P, Zaret KS 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125: 4909–4917 - PubMed
-
- Caravaca JM, Cano S, Gallego I, Daban JR 2005. Structural elements of bulk chromatin within metaphase chromosomes. Chromosome Res 13: 725–743 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
