Molecular paleontology: a biochemical model of the ancestral ribosome
- PMID: 23355613
- PMCID: PMC3597689
- DOI: 10.1093/nar/gkt023
Molecular paleontology: a biochemical model of the ancestral ribosome
Abstract
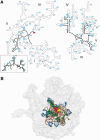
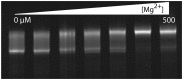
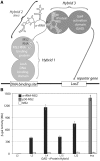
Ancient components of the ribosome, inferred from a consensus of previous work, were constructed in silico, in vitro and in vivo. The resulting model of the ancestral ribosome presented here incorporates ∼20% of the extant 23S rRNA and fragments of five ribosomal proteins. We test hypotheses that ancestral rRNA can: (i) assume canonical 23S rRNA-like secondary structure, (ii) assume canonical tertiary structure and (iii) form native complexes with ribosomal protein fragments. Footprinting experiments support formation of predicted secondary and tertiary structure. Gel shift, spectroscopic and yeast three-hybrid assays show specific interactions between ancestral rRNA and ribosomal protein fragments, independent of other, more recent, components of the ribosome. This robustness suggests that the catalytic core of the ribosome is an ancient construct that has survived billions of years of evolution without major changes in structure. Collectively, the data here support a model in which ancestors of the large and small subunits originated and evolved independently of each other, with autonomous functionalities.
Figures




Similar articles
-
Structure of the L1 protuberance in the ribosome.Nat Struct Biol. 2003 Feb;10(2):104-8. doi: 10.1038/nsb886. Nat Struct Biol. 2003. PMID: 12514741
-
The 23 S rRNA environment of ribosomal protein L9 in the 50 S ribosomal subunit.J Mol Biol. 2000 Apr 14;297(5):1129-43. doi: 10.1006/jmbi.2000.3621. J Mol Biol. 2000. PMID: 10764578
-
Effect of antibiotics on large ribosomal subunit assembly reveals possible function of 5 S rRNA.J Mol Biol. 1999 Sep 3;291(5):1025-34. doi: 10.1006/jmbi.1999.3030. J Mol Biol. 1999. PMID: 10518940
-
Progress toward an understanding of the structure and enzymatic mechanism of the large ribosomal subunit.Cold Spring Harb Symp Quant Biol. 2001;66:33-42. doi: 10.1101/sqb.2001.66.33. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762006 Review. No abstract available.
-
RNA-protein interactions in the Escherichia coli ribosome.Biochimie. 1991 Jul-Aug;73(7-8):927-36. doi: 10.1016/0300-9084(91)90134-m. Biochimie. 1991. PMID: 1720671 Review.
Cited by
-
What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention.Life (Basel). 2015 Jan 23;5(1):294-320. doi: 10.3390/life5010294. Life (Basel). 2015. PMID: 25625599 Free PMC article. Review.
-
Secondary structure and domain architecture of the 23S and 5S rRNAs.Nucleic Acids Res. 2013 Aug;41(15):7522-35. doi: 10.1093/nar/gkt513. Epub 2013 Jun 14. Nucleic Acids Res. 2013. PMID: 23771137 Free PMC article.
-
Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs.FEBS Open Bio. 2014 Feb 8;4:175-8. doi: 10.1016/j.fob.2014.01.010. eCollection 2014. FEBS Open Bio. 2014. PMID: 24649398 Free PMC article.
-
Mixed anhydrides at the intersection between peptide and RNA autocatalytic sets: evolution of biological coding.Interface Focus. 2023 Apr 14;13(3):20230009. doi: 10.1098/rsfs.2023.0009. eCollection 2023 Jun 6. Interface Focus. 2023. PMID: 37213924 Free PMC article.
-
Origins of tmRNA: the missing link in the birth of protein synthesis?Nucleic Acids Res. 2016 Sep 30;44(17):8041-51. doi: 10.1093/nar/gkw693. Epub 2016 Aug 2. Nucleic Acids Res. 2016. PMID: 27484476 Free PMC article. Review.
References
-
- Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC. Modeling a minimal ribosome based on comparative sequence analysis. J. Mol. Biol. 2002;321:215–234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

