Effects of DNA methylation on nucleosome stability
- PMID: 23355616
- PMCID: PMC3597673
- DOI: 10.1093/nar/gks893
Effects of DNA methylation on nucleosome stability
Abstract
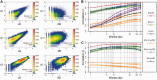
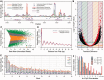
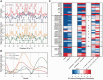
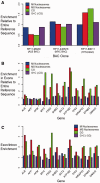
Methylation of DNA at CpG dinucleotides represents one of the most important epigenetic mechanisms involved in the control of gene expression in vertebrate cells. In this report, we conducted nucleosome reconstitution experiments in conjunction with high-throughput sequencing on 572 KB of human DNA and 668 KB of mouse DNA that was unmethylated or methylated in order to investigate the effects of this epigenetic modification on the positioning and stability of nucleosomes. The results demonstrated that a subset of nucleosomes positioned by nucleotide sequence was sensitive to methylation where the modification increased the affinity of these sequences for the histone octamer. The features that distinguished these nucleosomes from the bulk of the methylation-insensitive nucleosomes were an increase in the frequency of CpG dinucleotides and a unique rotational orientation of CpGs such that their minor grooves tended to face toward the histones in the nucleosome rather than away. These methylation-sensitive nucleosomes were preferentially associated with exons as compared to introns while unmethylated CpG islands near transcription start sites became enriched in nucleosomes upon methylation. The results of this study suggest that the effects of DNA methylation on nucleosome stability in vitro can recapitulate what has been observed in the cell and provide a direct link between DNA methylation and the structure and function of chromatin.
Figures


Similar articles
-
Links between DNA methylation and nucleosome occupancy in the human genome.Epigenetics Chromatin. 2017 Apr 11;10:18. doi: 10.1186/s13072-017-0125-5. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28413449 Free PMC article.
-
Contrasting chromatin organization of CpG islands and exons in the human genome.Genome Biol. 2010;11(7):R70. doi: 10.1186/gb-2010-11-7-r70. Epub 2010 Jul 5. Genome Biol. 2010. PMID: 20602769 Free PMC article.
-
A determining influence for CpG dinucleotides on nucleosome positioning in vitro.Nucleic Acids Res. 2004 Aug 13;32(14):4322-31. doi: 10.1093/nar/gkh749. Print 2004. Nucleic Acids Res. 2004. PMID: 15310836 Free PMC article.
-
DNA methylation, nucleosome formation and positioning.Brief Funct Genomic Proteomic. 2005 Feb;3(4):351-61. doi: 10.1093/bfgp/3.4.351. Brief Funct Genomic Proteomic. 2005. PMID: 15814025 Review.
-
Mechanisms of Inheritance of Chromatin States: From Yeast to Human.Annu Rev Biophys. 2025 May;54(1):59-79. doi: 10.1146/annurev-biophys-070524-091904. Epub 2024 Dec 23. Annu Rev Biophys. 2025. PMID: 39715046 Review.
Cited by
-
DNA methylation cues in nucleosome geometry, stability and unwrapping.Nucleic Acids Res. 2022 Feb 28;50(4):1864-1874. doi: 10.1093/nar/gkac097. Nucleic Acids Res. 2022. PMID: 35166834 Free PMC article.
-
IMPACTS OF DNA METHYLATION ON H2A.Z DEPOSITION AND NUCLEOSOME STABILITY.bioRxiv [Preprint]. 2025 Jul 31:2025.07.31.667981. doi: 10.1101/2025.07.31.667981. bioRxiv. 2025. PMID: 40766494 Free PMC article. Preprint.
-
Whole genome DNA methylation: beyond genes silencing.Oncotarget. 2017 Jan 17;8(3):5629-5637. doi: 10.18632/oncotarget.13562. Oncotarget. 2017. PMID: 27895318 Free PMC article. Review.
-
Transcriptional and epigenetic networks of helper T and innate lymphoid cells.Immunol Rev. 2014 Sep;261(1):23-49. doi: 10.1111/imr.12208. Immunol Rev. 2014. PMID: 25123275 Free PMC article. Review.
-
DNA methylation effects on tetra-nucleosome compaction and aggregation.Biophys J. 2014 Oct 7;107(7):1629-36. doi: 10.1016/j.bpj.2014.05.055. Biophys J. 2014. PMID: 25296315 Free PMC article.
References
-
- Feil R. Epigenetics: ready for the marks. Nature. 2009;461:359–360. - PubMed
-
- Lee J. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 2011;12:815–826. - PubMed
-
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. - PubMed
-
- Bird A. The Dinucleotide CG as a Genomic Signalling Module. J. Mol. Biol. 2011;409:47–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

