Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells
- PMID: 23356739
- PMCID: PMC3583671
- DOI: 10.1186/1471-2407-13-37
Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells
Abstract
Background: Post-translational modifications (PTMs) of histones and other proteins are perturbed in tumours. For example, reduced levels of acetylated H4K16 and trimethylated H4K20 are associated with high tumour grade and poor survival in breast cancer. Drug-like molecules that can reprogram selected histone PTMs in tumour cells are therefore of interest as potential cancer chemopreventive agents. In this study we assessed the effects of the phytocompounds garcinol and curcumin on histone and p53 modification in cancer cells, focussing on the breast tumour cell line MCF7.
Methods: Cell viability/proliferation assays, cell cycle analysis by flow cytometry, immunodetection of specific histone and p53 acetylation marks, western blotting, siRNA and RT-qPCR.
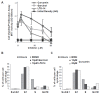
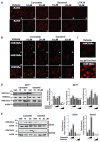
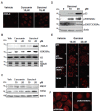
Results: Although treatment with curcumin, garcinol or the garcinol derivative LTK-14 hampered MCF7 cell proliferation, differential effects of these compounds on histone modifications were observed. Garcinol treatment resulted in a strong reduction in H3K18 acetylation, which is required for S phase progression. Similar effects of garcinol on H3K18 acetylation were observed in the osteosarcoma cells lines U2OS and SaOS2. In contrast, global levels of acetylated H4K16 and trimethylated H4K20 in MCF7 cells were elevated after garcinol treatment. This was accompanied by upregulation of DNA damage signalling markers such as γH2A.X, H3K56Ac, p53 and TIP60. In contrast, exposure of MCF7 cells to curcumin resulted in increased global levels of acetylated H3K18 and H4K16, and was less effective in inducing DNA damage markers. In addition to its effects on histone modifications, garcinol was found to block CBP/p300-mediated acetylation of the C-terminal activation domain of p53, but resulted in enhanced acetylation of p53K120, and accumulation of p53 in the cytoplasmic compartment. Finally, we show that the elevation of H4K20Me3 levels by garcinol correlated with increased expression of SUV420H2, and was prevented by siRNA targeting of SUV420H2.
Conclusion: In summary, although garcinol and curcumin can both inhibit histone acetyltransferase activities, our results show that these compounds have differential effects on cancer cells in culture. Garcinol treatment alters expression of chromatin modifying enzymes in MCF7 cells, resulting in reprogramming of key histone and p53 PTMs and growth arrest, underscoring its potential as a cancer chemopreventive agent.
Figures

Similar articles
-
Inhibitor of CBP Histone Acetyltransferase Downregulates p53 Activation and Facilitates Methylation at Lysine 27 on Histone H3.Molecules. 2018 Aug 2;23(8):1930. doi: 10.3390/molecules23081930. Molecules. 2018. PMID: 30072621 Free PMC article.
-
Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription.J Biol Chem. 2004 Dec 3;279(49):51163-71. doi: 10.1074/jbc.M409024200. Epub 2004 Sep 20. J Biol Chem. 2004. PMID: 15383533
-
Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair.J Biol Chem. 2017 Jul 14;292(28):11951-11959. doi: 10.1074/jbc.M117.788224. Epub 2017 May 25. J Biol Chem. 2017. PMID: 28546430 Free PMC article.
-
Histone modification therapy of cancer.Adv Genet. 2010;70:341-86. doi: 10.1016/B978-0-12-380866-0.60013-7. Adv Genet. 2010. PMID: 20920755 Review.
-
Role of histone acetylation in gastric cancer: implications of dietetic compounds and clinical perspectives.Epigenomics. 2019 Feb;11(3):349-362. doi: 10.2217/epi-2018-0081. Epub 2019 Jan 23. Epigenomics. 2019. PMID: 30672330 Review.
Cited by
-
Epi-nutrients for cancer prevention: Molecular mechanisms and emerging insights.Cell Biol Toxicol. 2025 Jul 15;41(1):116. doi: 10.1007/s10565-025-10054-2. Cell Biol Toxicol. 2025. PMID: 40663150 Free PMC article. Review.
-
Curcumin a potent cancer preventive agent: Mechanisms of cancer cell killing.Interv Med Appl Sci. 2014 Dec;6(4):139-46. doi: 10.1556/IMAS.6.2014.4.1. Epub 2014 Dec 22. Interv Med Appl Sci. 2014. PMID: 25598986 Free PMC article. Review.
-
Safety profile of 40% Garcinol from Garcinia indica in experimental rodents.Toxicol Rep. 2018 Jun 19;5:750-758. doi: 10.1016/j.toxrep.2018.06.009. eCollection 2018. Toxicol Rep. 2018. PMID: 29984188 Free PMC article.
-
Repurposing Plant-Based Histone Acetyltransferase Inhibitors: A Review of Novel Therapeutic Strategies Against Drug-Resistant Fungal Biofilms.Curr Microbiol. 2024 Nov 12;82(1):1. doi: 10.1007/s00284-024-03971-8. Curr Microbiol. 2024. PMID: 39532708 Review.
-
Curcumin and its Analogs and Carriers: Potential Therapeutic Strategies for Human Osteosarcoma.Int J Biol Sci. 2023 Feb 13;19(4):1241-1265. doi: 10.7150/ijbs.80590. eCollection 2023. Int J Biol Sci. 2023. PMID: 36923933 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

