Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation
- PMID: 23357851
- PMCID: PMC3641591
- DOI: 10.1038/cr.2013.16
Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation
Abstract
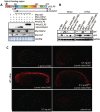
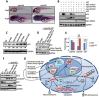
p53 protein turnover through the ubiquitination pathway is a vital mechanism in the regulation of its transcriptional activity; however, little is known about p53 turnover through proteasome-independent pathway(s). The digestive organ expansion factor (Def) protein is essential for the development of digestive organs. In zebrafish, loss of function of def selectively upregulates the expression of p53 response genes, which raises a question as to what is the relationship between Def and p53. We report here that Def is a nucleolar protein and that loss of function of def leads to the upregulation of p53 protein, which surprisingly accumulates in the nucleoli. Our extensive studies have demonstrated that Def can mediate the degradation of p53 protein and that this process is independent of the proteasome pathway, but dependent on the activity of Calpain3, a cysteine protease. Our findings define a novel nucleolar pathway that regulates the turnover function of p53, which will advance our understanding of p53's role in organogenesis and tumorigenesis.
Figures





Comment in
-
Proteasome-independent p53 degradation.Cell Res. 2013 May;23(5):597-8. doi: 10.1038/cr.2013.38. Epub 2013 Mar 12. Cell Res. 2013. PMID: 23478295 Free PMC article.
Similar articles
-
Regulation of the MDM2-p53 pathway by the nucleolar protein CSIG in response to nucleolar stress.Sci Rep. 2016 Nov 4;6:36171. doi: 10.1038/srep36171. Sci Rep. 2016. PMID: 27811966 Free PMC article.
-
Haploinsufficiency of Def activates p53-dependent TGFβ signalling and causes scar formation after partial hepatectomy.PLoS One. 2014 May 6;9(5):e96576. doi: 10.1371/journal.pone.0096576. eCollection 2014. PLoS One. 2014. PMID: 24801718 Free PMC article.
-
Proteasome-independent p53 degradation.Cell Res. 2013 May;23(5):597-8. doi: 10.1038/cr.2013.38. Epub 2013 Mar 12. Cell Res. 2013. PMID: 23478295 Free PMC article.
-
Nucleolus-localized Def-CAPN3 protein degradation pathway and its role in cell cycle control and ribosome biogenesis.J Genet Genomics. 2021 Nov 20;48(11):955-960. doi: 10.1016/j.jgg.2021.06.011. Epub 2021 Jul 10. J Genet Genomics. 2021. PMID: 34452850 Review.
-
The multiple levels of regulation by p53 ubiquitination.Cell Death Differ. 2010 Jan;17(1):86-92. doi: 10.1038/cdd.2009.77. Cell Death Differ. 2010. PMID: 19543236 Free PMC article. Review.
Cited by
-
Hepatic depletion of nucleolar protein mDEF causes excessive mitochondrial copper accumulation associated with p53 and NRF1 activation.iScience. 2023 Jun 26;26(7):107220. doi: 10.1016/j.isci.2023.107220. eCollection 2023 Jul 21. iScience. 2023. PMID: 37456842 Free PMC article.
-
Upf3a but not Upf1 mediates the genetic compensation response induced by leg1 deleterious mutations in an H3K4me3-independent manner.Cell Discov. 2023 Jun 27;9(1):63. doi: 10.1038/s41421-023-00550-2. Cell Discov. 2023. PMID: 37369707 Free PMC article.
-
Overexpression of PDGFRA cooperates with loss of NF1 and p53 to accelerate the molecular pathogenesis of malignant peripheral nerve sheath tumors.Oncogene. 2017 Feb 23;36(8):1058-1068. doi: 10.1038/onc.2016.269. Epub 2016 Aug 1. Oncogene. 2017. PMID: 27477693 Free PMC article.
-
p53 isoform Δ113p53/Δ133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage.Cell Res. 2015 Mar;25(3):351-69. doi: 10.1038/cr.2015.22. Epub 2015 Feb 20. Cell Res. 2015. PMID: 25698579 Free PMC article.
-
Loss of calpain3b in Zebrafish, a Model of Limb-Girdle Muscular Dystrophy, Increases Susceptibility to Muscle Defects Due to Elevated Muscle Activity.Genes (Basel). 2023 Feb 15;14(2):492. doi: 10.3390/genes14020492. Genes (Basel). 2023. PMID: 36833417 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

