Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome
- PMID: 23357950
- PMCID: PMC3554660
- DOI: 10.1371/journal.pone.0055105
Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome
Abstract
Background: Mirid plant bugs (Hemiptera: Miridae) are economically important insect pests of many crops worldwide. The western tarnished plant bug Lygus hesperus Knight is a pest of cotton, alfalfa, fruit and vegetable crops, and potentially of several emerging biofuel and natural product feedstocks in the western US. However, little is known about the underlying molecular genetics, biochemistry, or physiology of L. hesperus, including their ability to survive extreme environmental conditions.
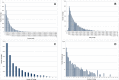
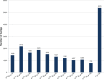
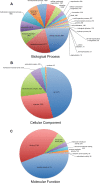
Methodology/principal findings: We used 454 pyrosequencing of a normalized adult cDNA library and de novo assembly to obtain an adult L. hesperus transcriptome consisting of 1,429,818 transcriptomic reads representing 36,131 transcript isoforms (isotigs) that correspond to 19,742 genes. A search of the transcriptome against deposited L. hesperus protein sequences revealed that 86 out of 87 were represented. Comparison with the non-redundant database indicated that 54% of the transcriptome exhibited similarity (e-value ≤ 1(-5)) with known proteins. In addition, Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations, and potential Pfam domains were assigned to each transcript isoform. To gain insight into the molecular basis of the L. hesperus thermal stress response we used transcriptomic sequences to identify 52 potential heat shock protein (Hsp) homologs. A subset of these transcripts was sequence verified and their expression response to thermal stress monitored by semi-quantitative PCR. Potential homologs of Hsp70, Hsp40, and 2 small Hsps were found to be upregulated in the heat-challenged adults, suggesting a role in thermotolerance.
Conclusions/significance: The L. hesperus transcriptome advances the underlying molecular understanding of this arthropod pest by significantly increasing the number of known genes, and provides the basis for further exploration and understanding of the fundamental mechanisms of abiotic stress responses.
Conflict of interest statement
Figures






References
-
- Beards GW, Strong FE (1966) Photoperiod in relation to diapause in Lygus hesperus Knight. Hilgardia 37: 345–362.
-
- Leigh TF (1966) A reproductive diapause in Lygus hesperus Knight. J Econ Entomol 59: 1280–1281.
-
- Spurgeon DW, Brent CS (2010) Morphological characters of diapause in Lygus hesperus Knight (Hemiptera: Miridae). J Entomol Sci 45: 303–316.
-
- Kelton LA (1975) The Lygus Bugs (Genus Lygus Hahn) of North America (Heteroptera: Miridae). Mem Entomol Soc Can 95: 1–101.
-
- Scott DR (1977) An annotated listing of host plants of Lygus hesperus Knight. Entomo Soc Am Bull 23: 19–22.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

