Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response
- PMID: 23358328
- PMCID: PMC3554634
- DOI: 10.1371/journal.ppat.1003141
Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response
Abstract
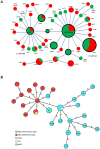
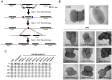
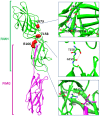
Adherent-invasive Escherichia coli (AIEC) are abnormally predominant on Crohn's disease (CD) ileal mucosa. AIEC reference strain LF82 adheres to ileal enterocytes via the common type 1 pili adhesin FimH and recognizes CEACAM6 receptors abnormally expressed on CD ileal epithelial cells. The fimH genes of 45 AIEC and 47 non-AIEC strains were sequenced. The phylogenetic tree based on fimH DNA sequences indicated that AIEC strains predominantly express FimH with amino acid mutations of a recent evolutionary origin - a typical signature of pathoadaptive changes of bacterial pathogens. Point mutations in FimH, some of a unique AIEC-associated nature, confer AIEC bacteria a significantly higher ability to adhere to CEACAM-expressing T84 intestinal epithelial cells. Moreover, in the LF82 strain, the replacement of fimH(LF82) (expressing FimH with an AIEC-associated mutation) with fimH(K12) (expressing FimH of commensal E. coli K12) decreased the ability of bacteria to persist and to induce severe colitis and gut inflammation in infected CEABAC10 transgenic mice expressing human CEACAM receptors. Our results highlight a mechanism of AIEC virulence evolution that involves selection of amino acid mutations in the common bacterial traits, such as FimH protein, and leads to the development of chronic inflammatory bowel disease (IBD) in a genetically susceptible host. The analysis of fimH SNPs may be a useful method to predict the potential virulence of E. coli isolated from IBD patients for diagnostic or epidemiological studies and to identify new strategies for therapeutic intervention to block the interaction between AIEC and gut mucosa in the early stages of IBD.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Xavier RJ, Podolsky DK (2007) Unravelling the pathogenesis of inflammatory bowel disease. Nature 448: 427–434. - PubMed
-
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127: 412–421. - PubMed
-
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, et al. (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115: 1405–1413. - PubMed
-
- Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, et al. (2008) Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol 298: 397–409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

