Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies
- PMID: 23359068
- PMCID: PMC3570098
- DOI: 10.1084/jem.20121977
Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies
Abstract
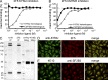
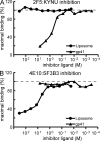
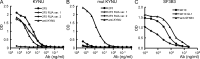
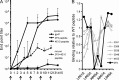
Many human monoclonal antibodies that neutralize multiple clades of HIV-1 are polyreactive and bind avidly to mammalian autoantigens. Indeed, the generation of neutralizing antibodies to the 2F5 and 4E10 epitopes of HIV-1 gp41 in man may be proscribed by immune tolerance because mice expressing the V(H) and V(L) regions of 2F5 have a block in B cell development that is characteristic of central tolerance. This developmental blockade implies the presence of tolerizing autoantigens that are mimicked by the membrane-proximal external region of HIV-1 gp41. We identify human kynureninase (KYNU) and splicing factor 3b subunit 3 (SF3B3) as the primary conserved, vertebrate self-antigens recognized by the 2F5 and 4E10 antibodies, respectively. 2F5 binds the H4 domain of KYNU which contains the complete 2F5 linear epitope (ELDKWA). 4E10 recognizes an epitope of SF3B3 that is strongly dependent on hydrophobic interactions. Opossums carry a rare KYNU H4 domain that abolishes 2F5 binding, but they retain the SF3B3 4E10 epitope. Immunization of opossums with HIV-1 gp140 induced extraordinary titers of serum antibody to the 2F5 ELDKWA epitope but little or nothing to the 4E10 determinant. Identification of structural motifs shared by vertebrates and HIV-1 provides direct evidence that immunological tolerance can impair humoral responses to HIV-1.
Figures





References
-
- Alam S.M., McAdams M., Boren D., Rak M., Scearce R.M., Gao F., Camacho Z.T., Gewirth D., Kelsoe G., Chen P., Haynes B.F. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424–4435 - PMC - PubMed
-
- Alam S.M., Scearce R.M., Parks R.J., Plonk K., Plonk S.G., Sutherland L.L., Gorny M.K., Zolla-Pazner S., Vanleeuwen S., Moody M.A., et al. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82:115–125 10.1128/JVI.00927-07 - DOI - PMC - PubMed
-
- Alam S.M., Morelli M., Dennison S.M., Liao H.X., Zhang R., Xia S.M., Rits-Volloch S., Sun L., Harrison S.C., Haynes B.F., Chen B. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA. 106:20234–20239 10.1073/pnas.0908713106 - DOI - PMC - PubMed
-
- Alam S.M., Liao H.X., Dennison S.M., Jaeger F., Parks R., Anasti K., Foulger A., Donathan M., Lucas J., Verkoczy L., et al. 2011. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J. Virol. 85:11725–11731 10.1128/JVI.05680-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

