Targeting the RAS oncogene
- PMID: 23360111
- PMCID: PMC3804031
- DOI: 10.1517/14728222.2013.764990
Targeting the RAS oncogene
Abstract
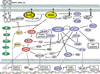
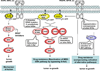
Introduction: The Ras proteins (K-Ras, N-Ras, and H-Ras) are GTPases that function as molecular switches for a variety of critical cellular activities and their function is tightly and temporally regulated in normal cells. Oncogenic mutations in the RAS genes, which create constitutively-active Ras proteins, can result in uncontrolled proliferation or survival in tumor cells.
Areas covered: The paper discusses three therapeutic approaches targeting the Ras pathway in cancer: i) Ras itself, ii) Ras downstream pathways, and iii) synthetic lethality. The most adopted approach is targeting Ras downstream signaling, and specifically the PI3K-AKT-mTOR and Raf-MEK pathways, as they are frequently major oncogenic drivers in cancers with high Ras signaling. Although direct targeting of Ras has not been successful clinically, newer approaches being investigated in preclinical studies, such as RNA interference-based and synthetic lethal approaches, promise great potential for clinical application.
Expert opinion: The challenges of current and emerging therapeutics include the lack of "tumor specificity" and their limitation to those cancers which are "dependent" on aberrant Ras signaling for survival. While the newer approaches have the potential to overcome these limitations, they also highlight the importance of robust preclinical studies and bidirectional translational research for successful clinical development of Ras-related targeted therapies.
Similar articles
-
Approaches to Ras signaling modulation and treatment of Ras-dependent disorders: a patent review (2007--present).Expert Opin Ther Pat. 2012 Nov;22(11):1263-87. doi: 10.1517/13543776.2012.728586. Epub 2012 Sep 26. Expert Opin Ther Pat. 2012. PMID: 23009088 Review.
-
Targeting the RAS upstream and downstream signaling pathway for cancer treatment.Eur J Pharmacol. 2024 Sep 15;979:176727. doi: 10.1016/j.ejphar.2024.176727. Epub 2024 Jun 10. Eur J Pharmacol. 2024. PMID: 38866361 Review.
-
Improving Prospects for Targeting RAS.J Clin Oncol. 2015 Nov 1;33(31):3650-9. doi: 10.1200/JCO.2015.62.1052. Epub 2015 Sep 14. J Clin Oncol. 2015. PMID: 26371146 Review.
-
The greedy nature of mutant RAS: a boon for drug discovery targeting cancer metabolism?Acta Biochim Biophys Sin (Shanghai). 2016 Jan;48(1):17-26. doi: 10.1093/abbs/gmv102. Epub 2015 Oct 19. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26487443 Review.
-
Past, Present, and Future of Targeting Ras for Cancer Therapies.Mini Rev Med Chem. 2016;16(5):345-57. doi: 10.2174/1389557515666151001154111. Mini Rev Med Chem. 2016. PMID: 26423695 Review.
Cited by
-
Copper bioavailability is a KRAS-specific vulnerability in colorectal cancer.Nat Commun. 2020 Jul 24;11(1):3701. doi: 10.1038/s41467-020-17549-y. Nat Commun. 2020. PMID: 32709883 Free PMC article.
-
Pancreatic Cancer from Molecular Pathways to Treatment Opinion.J Cancer. 2016 Jun 25;7(10):1328-39. doi: 10.7150/jca.15419. eCollection 2016. J Cancer. 2016. PMID: 27390608 Free PMC article. Review.
-
Protein kinase C-δ inactivation inhibits the proliferation and survival of cancer stem cells in culture and in vivo.BMC Cancer. 2014 Feb 14;14:90. doi: 10.1186/1471-2407-14-90. BMC Cancer. 2014. PMID: 24528676 Free PMC article.
-
RAS signaling and anti-RAS therapy: lessons learned from genetically engineered mouse models, human cancer cells, and patient-related studies.Acta Biochim Biophys Sin (Shanghai). 2016 Jan;48(1):27-38. doi: 10.1093/abbs/gmv090. Epub 2015 Sep 7. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26350096 Free PMC article. Review.
-
KRAS and HRAS mutations confer resistance to MET targeting in preclinical models of MET-expressing tumor cells.Mol Oncol. 2015 Aug;9(7):1434-46. doi: 10.1016/j.molonc.2015.04.001. Epub 2015 Apr 14. Mol Oncol. 2015. PMID: 25933688 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous