RNA-guided editing of bacterial genomes using CRISPR-Cas systems
- PMID: 23360965
- PMCID: PMC3748948
- DOI: 10.1038/nbt.2508
RNA-guided editing of bacterial genomes using CRISPR-Cas systems
Abstract
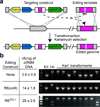
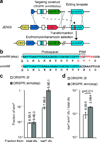
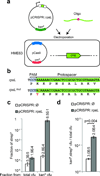
Here we use the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated Cas9 endonuclease complexed with dual-RNAs to introduce precise mutations in the genomes of Streptococcus pneumoniae and Escherichia coli. The approach relies on dual-RNA:Cas9-directed cleavage at the targeted genomic site to kill unmutated cells and circumvents the need for selectable markers or counter-selection systems. We reprogram dual-RNA:Cas9 specificity by changing the sequence of short CRISPR RNA (crRNA) to make single- and multinucleotide changes carried on editing templates. Simultaneous use of two crRNAs enables multiplex mutagenesis. In S. pneumoniae, nearly 100% of cells that were recovered using our approach contained the desired mutation, and in E. coli, 65% that were recovered contained the mutation, when the approach was used in combination with recombineering. We exhaustively analyze dual-RNA:Cas9 target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements, suggesting the versatility of this technique for bacterial genome engineering.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures


References
-
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. - PubMed
-
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
-
- Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. - PubMed
-
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

