A point mutation in Semaphorin 4A associates with defective endosomal sorting and causes retinal degeneration
- PMID: 23360997
- PMCID: PMC3562463
- DOI: 10.1038/ncomms2420
A point mutation in Semaphorin 4A associates with defective endosomal sorting and causes retinal degeneration
Abstract
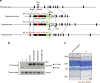
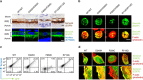
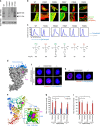
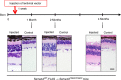
Semaphorin 4A (Sema4A) has an essential role in photoreceptor survival. In humans, mutations in Sema4A are thought to contribute to retinal degenerative diseases. Here we generate a series of knock-in mouse lines with corresponding mutations (D345H, F350C or R713Q) in the Sema4A gene and find that Sema4A(F350C) causes retinal degeneration phenotypes. The F350C mutation results in abnormal localization of the Sema4A protein, leading to impaired endosomal sorting of molecules indispensable for photoreceptor survival. Additionally, protein structural modelling reveals that the side chain of the 350th amino acid is critical to retain the proper protein conformation. Furthermore, Sema4A gene transfer successfully prevents photoreceptor degeneration in Sema4A(F350C/F350C) and Sema4A(-/-) mice. Thus, our findings not only indicate the importance of the Sema4A protein conformation in human and mouse retina homeostasis but also identify a novel therapeutic target for retinal degenerative diseases.
Figures



References
-
- Pacione L. R. et al. Progress toward understanding the genetic and biochemical mechanisms of inherited photoreceptor degenerations. Annu. Rev. Neurosci. 26, 657–700 (2003) . - PubMed
-
- Wright A. F. et al. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 11, 273–284 (2010) . - PubMed
-
- Kolodkin A. L., Matthes D. J. & Goodman C. S.. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 75, 1389–1399 (1993) . - PubMed
-
- Serini G. et al. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature 424, 391–397 (2003) . - PubMed
-
- Neufeld G. & Kessler O.. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer 8, 632–645 (2008) . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

