Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53
- PMID: 23360998
- PMCID: PMC3562459
- DOI: 10.1038/ncomms2361
Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53
Abstract
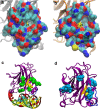
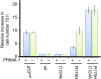
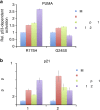
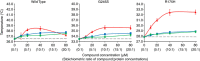
The tumour suppressor p53 is the most frequently mutated gene in human cancer. Reactivation of mutant p53 by small molecules is an exciting potential cancer therapy. Although several compounds restore wild-type function to mutant p53, their binding sites and mechanisms of action are elusive. Here computational methods identify a transiently open binding pocket between loop L1 and sheet S3 of the p53 core domain. Mutation of residue Cys124, located at the centre of the pocket, abolishes p53 reactivation of mutant R175H by PRIMA-1, a known reactivation compound. Ensemble-based virtual screening against this newly revealed pocket selects stictic acid as a potential p53 reactivation compound. In human osteosarcoma cells, stictic acid exhibits dose-dependent reactivation of p21 expression for mutant R175H more strongly than does PRIMA-1. These results indicate the L1/S3 pocket as a target for pharmaceutical reactivation of p53 mutants.
Figures



References
-
- Olivier M. et al. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19, 607–614 (2002) . - PubMed
-
- Soussi T. & Beroud C.. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 1, 233–240 (2001) . - PubMed
-
- Martins C. P., Brown-Swigart L. & Evan G. I.. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334 (2006) . - PubMed
-
- Selivanova G. & Wiman K. G.. Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene 26, 2243–2254 (2007) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R42 AI066758/AI/NIAID NIH HHS/United States
- R42AI066758/AI/NIAID NIH HHS/United States
- T32 CA009054/CA/NCI NIH HHS/United States
- R01 AI078000/AI/NIAID NIH HHS/United States
- F30 CA174329/CA/NCI NIH HHS/United States
- R01 CA112560/CA/NCI NIH HHS/United States
- DP2 OD007237/OD/NIH HHS/United States
- T32CA009054/CA/NCI NIH HHS/United States
- R01CA112560/CA/NCI NIH HHS/United States
- R01AI78000/AI/NIAID NIH HHS/United States
- T15LM07443/LM/NLM NIH HHS/United States
- DP2-OD007237/OD/NIH HHS/United States
- T15 LM007443/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

