Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals
- PMID: 23361376
- PMCID: PMC3618476
- DOI: 10.1039/c3cs35458j
Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals
Abstract
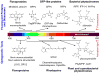
Near-infrared light is favourable for imaging in mammalian tissues due to low absorbance of hemoglobin, melanin, and water. Therefore, fluorescent proteins, biosensors and optogenetic constructs for optimal imaging, optical readout and light manipulation in mammals should have fluorescence and action spectra within the near-infrared window. Interestingly, natural Bacterial Phytochrome Photoreceptors (BphPs) utilize the low molecular weight biliverdin, found in most mammalian tissues, as a photoreactive chromophore. Due to their near-infrared absorbance BphPs are preferred templates for designing optical molecular tools for applications in mammals. Moreover, BphPs spectrally complement existing genetically-encoded probes. Several BphPs were already developed into the near-infrared fluorescent variants. Based on the analysis of the photochemistry and structure of BphPs we suggest a variety of possible BphP-based fluorescent proteins, biosensors, and optogenetic tools. Putative design strategies and experimental considerations for such probes are discussed.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

