HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings
- PMID: 23362207
- PMCID: PMC3584530
- DOI: 10.1105/tpc.112.096313
HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings
Abstract
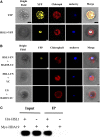
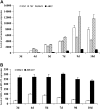
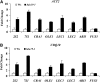
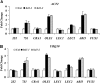
The seed maturation genes are specifically and highly expressed during late embryogenesis. In this work, yeast two-hybrid, bimolecular fluorescence complementation, and coimmunoprecipitation assays revealed that HISTONE DEACETYLASE19 (HDA19) interacted with the HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE2-LIKE1 (HSL1), and the zinc-finger CW [conserved Cys (C) and Trp (W) residues] domain of HSL1 was responsible for the interaction. Furthermore, we found that mutations in HDA19 resulted in the ectopic expression of seed maturation genes in seedlings, which was associated with increased levels of gene activation marks, such as Histone H3 acetylation (H3ac), Histone H4 acetylation (H4ac), and Histone H3 Lys 4 tri-methylation (H3K4me3), but decreased levels of the gene repression mark Histone H3 Lys 27 tri-methylation (H3K27me3) in the promoter and/or coding regions. In addition, elevated transcription of certain seed maturation genes was also found in the hsl1 mutant seedlings, which was also accompanied by the enrichment of gene activation marks but decreased levels of the gene repression mark. Chromatin immunoprecipitation assays showed that HDA19 could directly bind to the chromatin of the seed maturation genes. These results suggest that HDA19 and HSL1 may act together to repress seed maturation gene expression during germination. Further genetic analyses revealed that the homozygous hsl1 hda19 double mutants are embryonic lethal, suggesting that HDA19 and HSL1 may play a vital role during embryogenesis.
Figures





Similar articles
-
Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation.Plant Cell. 2013 Jan;25(1):149-66. doi: 10.1105/tpc.112.108191. Epub 2013 Jan 31. Plant Cell. 2013. PMID: 23371947 Free PMC article.
-
HSI2 Repressor Recruits MED13 and HDA6 to Down-Regulate Seed Maturation Gene Expression Directly During Arabidopsis Early Seedling Growth.Plant Cell Physiol. 2016 Aug;57(8):1689-706. doi: 10.1093/pcp/pcw095. Epub 2016 May 11. Plant Cell Physiol. 2016. PMID: 27335347
-
SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme.Nat Commun. 2015 Jul 1;6:7243. doi: 10.1038/ncomms8243. Nat Commun. 2015. PMID: 26129778 Free PMC article.
-
Decoding histone 3 lysine methylation: Insights into seed germination and flowering.Curr Opin Plant Biol. 2024 Oct;81:102598. doi: 10.1016/j.pbi.2024.102598. Epub 2024 Jul 9. Curr Opin Plant Biol. 2024. PMID: 38986392 Review.
-
Genetic activity during early plant embryogenesis.Biochem J. 2020 Oct 16;477(19):3743-3767. doi: 10.1042/BCJ20190161. Biochem J. 2020. PMID: 33045058 Free PMC article. Review.
Cited by
-
HISTONE DEACETYLASE19 Controls Ovule Number Determination and Transmitting Tract Differentiation.Plant Physiol. 2024 Mar 29;194(4):2117-2135. doi: 10.1093/plphys/kiad629. Plant Physiol. 2024. PMID: 38060625 Free PMC article.
-
Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants.Plant Commun. 2021 Nov 26;3(1):100267. doi: 10.1016/j.xplc.2021.100267. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059633 Free PMC article. Review.
-
POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis.Proc Natl Acad Sci U S A. 2016 Dec 20;113(51):14858-14863. doi: 10.1073/pnas.1618618114. Epub 2016 Dec 5. Proc Natl Acad Sci U S A. 2016. PMID: 27930340 Free PMC article.
-
VAL genes regulate vegetative phase change via miR156-dependent and independent mechanisms.PLoS Genet. 2021 Jun 28;17(6):e1009626. doi: 10.1371/journal.pgen.1009626. eCollection 2021 Jun. PLoS Genet. 2021. PMID: 34181637 Free PMC article.
-
Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression.Genes (Basel). 2023 May 30;14(6):1199. doi: 10.3390/genes14061199. Genes (Basel). 2023. PMID: 37372378 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

