HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes
- PMID: 23362208
- PMCID: PMC3584540
- DOI: 10.1105/tpc.112.107045
HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes
Abstract
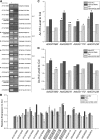
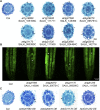
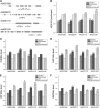
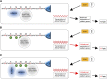
The differentiation of hair (H) and non-hair (N) cells in the Arabidopsis thaliana root epidermis is dependent on positional relationships with underlying cortical cells. We previously found that histone acetylation relays positional information and that a mutant altered in the histone deacetylase gene family member HISTONE DEACETYLASE 18 (HDA18) exhibits altered H and N epidermal cell patterning. Here, we report that HDA18 has in vitro histone deacetylase activity and that both mutation and overexpression of HDA18 led to cells at the N position having H fate. The HDA18 protein physically interacted with histones related to a specific group of kinase genes, which are demonstrated in this study to be components of a positional information relay system. Both down- and upregulation of HDA18 increased transcription of the targeted kinase genes. Interestingly, the acetylation levels of histone 3 lysine 9 (H3K9), histone 3 lysine 14 (H3K14) and histone 3 lysine 18 (H3K18) at the kinase genes were differentially affected by down- or upregulation of HDA18, which explains why the transcription levels of the four HDA18-target kinase genes increased in all lines with altered HDA18 expression. Our results reveal the surprisingly complex mechanism by which HDA18 affects cellular patterning in Arabidopsis root epidermis.
Figures



Comment in
-
Trust in nature.Plant Signal Behav. 2013 May;8(5):e23936. doi: 10.4161/psb.23936. Epub 2013 Feb 20. Plant Signal Behav. 2013. PMID: 23425922 Free PMC article.
References
-
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 - PubMed
-
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
-
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 - PubMed
-
- Caro E., Castellano M.M., Gutierrez C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

