Bacterial small RNA-based negative regulation: Hfq and its accomplices
- PMID: 23362267
- PMCID: PMC3605619
- DOI: 10.1074/jbc.R112.441386
Bacterial small RNA-based negative regulation: Hfq and its accomplices
Abstract
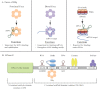
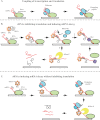
A large group of bacterial small regulatory RNAs (sRNAs) use the Hfq chaperone to mediate pairing with and regulation of mRNAs. Recent findings help to clarify how Hfq acts and highlight the role of the endonuclease RNase E and its associated proteins (the degradosome) in negative regulation by these sRNAs. sRNAs frequently uncouple transcription and translation by blocking ribosome access to the mRNA, allowing other proteins access to the mRNA. As more examples of sRNA-mediated regulation are studied, more variations on how Hfq, RNase E, and other proteins collaborate to bring about sRNA-based regulation are being found.
Figures
References
-
- Jacob F., Monod J. (1961) Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 - PubMed
-
- Gottesman S. (2011) Roles of mRNA stability, translational regulation, and small RNAs in stress response regulation. in Bacterial Stress Responses (Storz G., Hengge R. eds) 2nd Ed., pp. 59–73, ASM Press, Washington, D.C.
-
- Fröhlich K. S., Vogel J. (2009) Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12, 674–682 - PubMed
-
- Carmichael G. G., Weber K., Niveleau A., Wahba A. J. (1975) The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J. Biol. Chem. 250, 3607–3612 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

