Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer
- PMID: 23362326
- PMCID: PMC3596045
- DOI: 10.1158/1078-0432.CCR-12-2726
Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer
Abstract
Purpose: Endometriosis, a largely benign, chronic inflammatory disease, is an independent risk factor for endometrioid and clear cell epithelial ovarian tumors. We aimed to identify plasma miRNAs that can be used to differentiate patients with endometriosis and ovarian cancer from healthy individuals.
Experimental design: We conducted a two-stage exploratory study to investigate the use of plasma miRNA profiling to differentiate between patients with endometriosis, patients with endometriosis-associated ovarian cancer (EAOC), and healthy individuals. In the first stage, using global profiling of more than 1,000 miRNAs via reverse transcriptase quantitative PCR (RT-qPCR) in a 20-patient initial screening cohort, we identified 23 candidate miRNAs, which are differentially expressed between healthy controls (n = 6), patients with endometriosis (n = 7), and patients with EAOC (n = 7) based on the fold changes. In the second stage, the 23 miRNAs were further tested in an expanded cohort (n = 88) of healthy individuals (n = 20), endometriosis (n = 33), EAOC (n = 14), and serous ovarian cancer cases (SOC; n = 21, included as controls).
Results: We identified three distinct miRNA signatures with reliable differential expression between healthy individuals, patients with endometriosis, and patients with EAOC. When profiled against the control SOC category, our results revealed different miRNAs, suggesting that the identified signatures are reflective of disease-specific pathogenic mechanisms. This was further supported by the fact that the majority of miRNAs differentially expressed in human EAOCs were mirrored in a double transgenic mouse EAOC model.
Conclusion: Our study reports for the first time that distinct plasma miRNA expression patterns may serve as highly specific and sensitive diagnostic biomarkers to discriminate between healthy, endometriosis, and EAOC cases.
©2013 AACR.
Conflict of interest statement
Figures

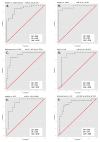

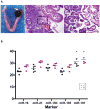

Comment in
-
Plasma microRNAs in ovarian cancer--response.Clin Cancer Res. 2013 Jun 15;19(12):3326. doi: 10.1158/1078-0432.CCR-13-1022. Epub 2013 Apr 24. Clin Cancer Res. 2013. PMID: 23616635 Free PMC article. No abstract available.
-
Plasma microRNAs in ovarian cancer--letter.Clin Cancer Res. 2013 Jun 15;19(12):3325. doi: 10.1158/1078-0432.CCR-13-0561. Epub 2013 Apr 24. Clin Cancer Res. 2013. PMID: 23616637 No abstract available.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Clarke-Pearson DL. Screening for Ovarian Cancer. N Engl J Med. 2009;361:170–7. - PubMed
-
- Engel J, Eckel R, Schubert-Fritschle G, Kerr J, Kuhn W, Diebold J, et al. Moderate progress for ovarian cancer in the last 20 years: prolongation of survival, but no improvement in the cure rate. Eur J Cancer. 2002;38:2435–45. - PubMed
-
- Bast RC, Jr, Badgwell D, Lu Z, Marquez R, Rosen D, Liu J, et al. New tumor markers: CA125 and beyond. Int J Gynecol Cancer. 2005;15:274–81. - PubMed
-
- Gagnon A, Ye B. Discovery and application of protein biomarkers for ovarian cancer. Curr Opin Obstet Gynecol. 2008;20:9–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

